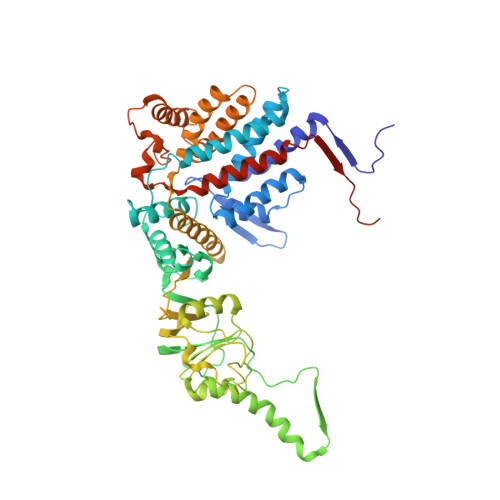
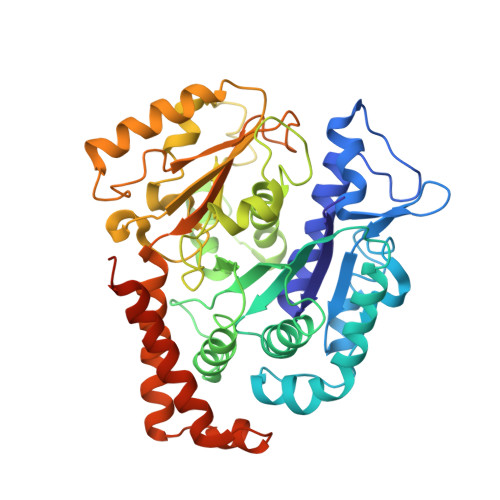
Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Liu, C., Jin, M., Wang, S., Han, W., Zhao, Q., Wang, Y., Xu, C., Diao, L., Yin, Y., Peng, C., Bao, L., Wang, Y., Cong, Y.(2023) Commun Biol 6: 531-531
- PubMed: 37193829
- DOI: https://doi.org/10.1038/s42003-023-04915-x
- Primary Citation of Related Structures:
7WZ3, 7X0A, 7X0S, 7X0V, 7X3J, 7X3U, 7X6Q, 7X7Y - PubMed Abstract:
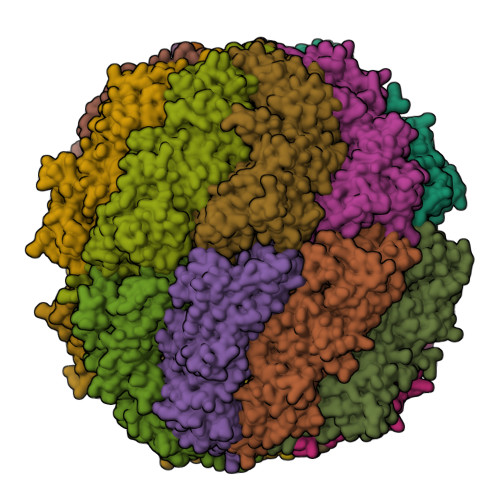
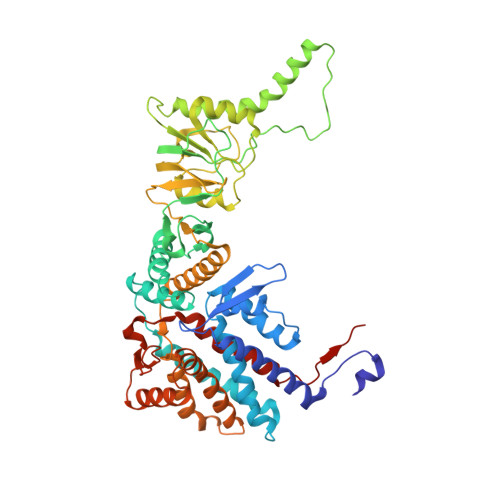
The eukaryotic chaperonin TRiC/CCT assists the folding of about 10% of cytosolic proteins through an ATP-driven conformational cycle, and the essential cytoskeleton protein tubulin is the obligate substrate of TRiC. Here, we present an ensemble of cryo-EM structures of endogenous human TRiC throughout its ATPase cycle, with three of them revealing endogenously engaged tubulin in different folding stages. The open-state TRiC-tubulin-S1 and -S2 maps show extra density corresponding to tubulin in the cis-ring chamber of TRiC. Our structural and XL-MS analyses suggest a gradual upward translocation and stabilization of tubulin within the TRiC chamber accompanying TRiC ring closure. In the closed TRiC-tubulin-S3 map, we capture a near-natively folded tubulin-with the tubulin engaging through its N and C domains mainly with the A and I domains of the CCT3/6/8 subunits through electrostatic and hydrophilic interactions. Moreover, we also show the potential role of TRiC C-terminal tails in substrate stabilization and folding. Our study delineates the pathway and molecular mechanism of TRiC-mediated folding of tubulin along the ATPase cycle of TRiC, and may also inform the design of therapeutic agents targeting TRiC-tubulin interactions.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, 200031, Shanghai, China.