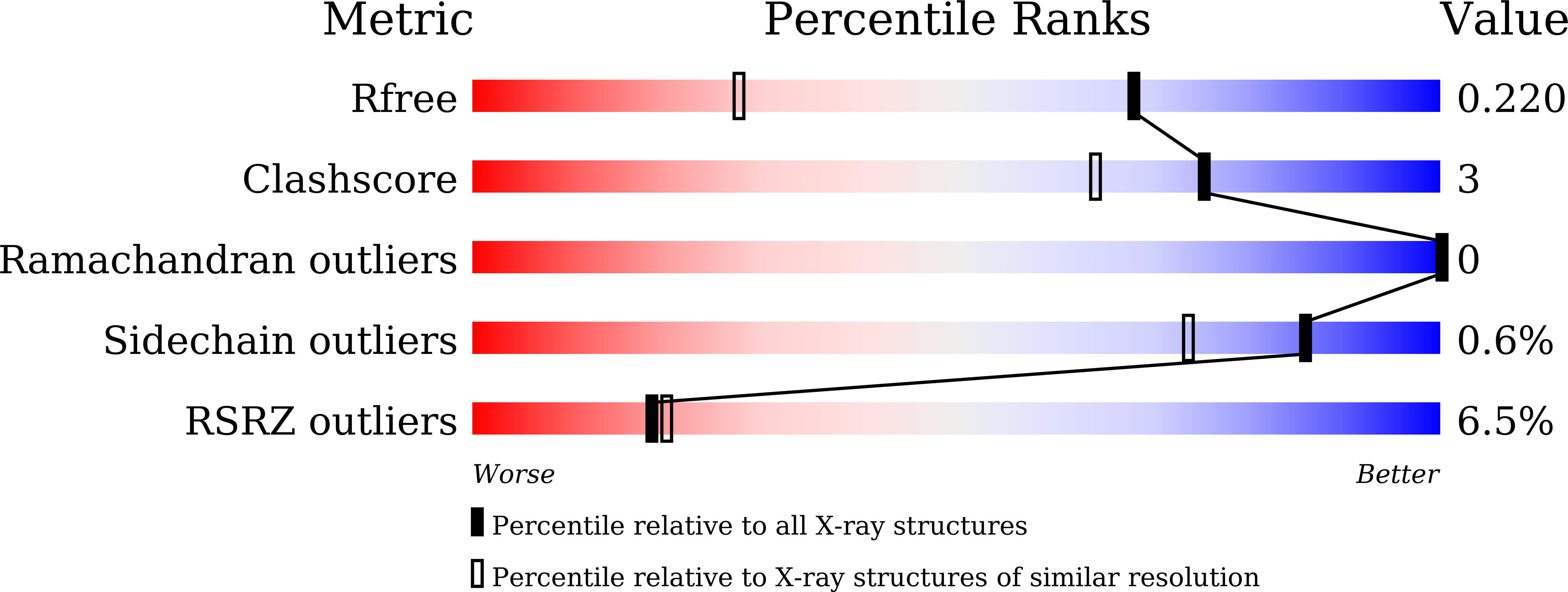
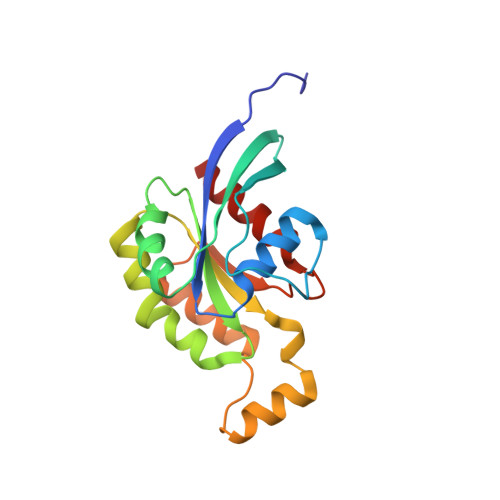
A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
Jiang, H., Zu, S., Lu, Y., Sun, Z., Adeerjiang, A., Guo, Q., Zhang, H., Dong, C., Wu, Q., Ding, H., Du, D., Wang, M., Liu, C., Tang, Y., Liang, Z., Luo, C.(2023) J Struct Biol 215: 107942-107942
- PubMed: 36781028
- DOI: https://doi.org/10.1016/j.jsb.2023.107942
- Primary Citation of Related Structures:
7WZA, 7WZC - PubMed Abstract:
Small GTPase RhoA switches from GTP-bound state to GDP-bound state by hydrolyzing GTP, which is accelerated by GTPases activating proteins (GAPs). However, less study of RhoA structural dynamic changes was conducted during this process, which is essential for understanding the molecular mechanism of GAP dissociation. Here, we solved a RhoA structure in GDP-bound state with switch II flipped outward. Because lacking the intermolecular interactions with guanine nucleotide, we proposed this conformation of RhoA could be an intermediate after GAP dissociation. Further molecular dynamics simulations found the conformational changes of switch regions are indeed existing in RhoA and involved in the regulation of GAP dissociation and GEF recognition. Besides, the guanine nucleotide binding pocket extended to switch II region, indicating a potential "druggable" cavity for RhoA. Taken together, our study provides a deeper understanding of the dynamic properties of RhoA switch regions and highlights the direction for future drug development.
Organizational Affiliation:
School of Pharmaceutical Science and Technology, Hangzhou Institute for Advanced Study, UCAS, Hangzhou 310024, China; State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, China; University of Chinese Academy of Sciences (UCAS), 19 Yuquan Road, Beijing 100049, China.