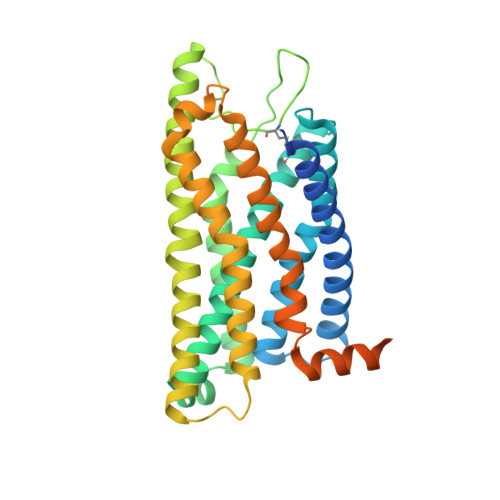
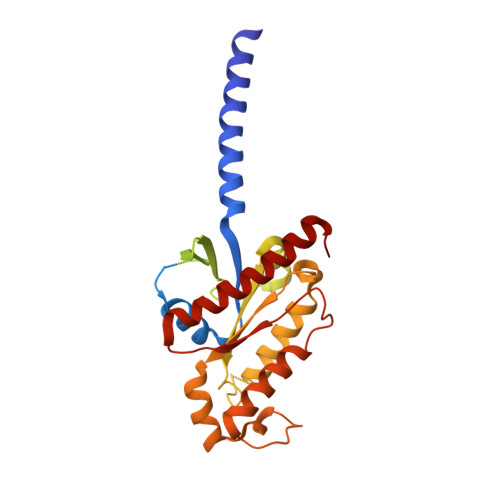
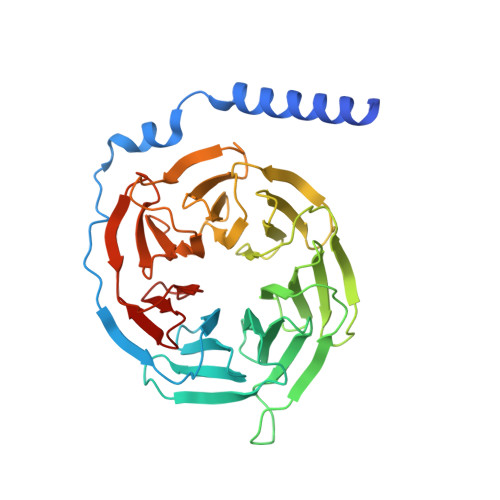
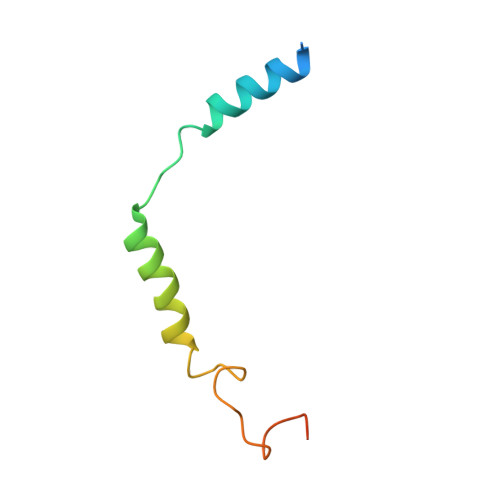
Structural basis of FPR2 in recognition of A beta 42 and neuroprotection by humanin.
Zhu, Y., Lin, X., Zong, X., Han, S., Wang, M., Su, Y., Ma, L., Chu, X., Yi, C., Zhao, Q., Wu, B.(2022) Nat Commun 13: 1775-1775
- PubMed: 35365641
- DOI: https://doi.org/10.1038/s41467-022-29361-x
- Primary Citation of Related Structures:
7WVU, 7WVV, 7WVW, 7WVX, 7WVY - PubMed Abstract:
Formyl peptide receptor 2 (FPR2) has been shown to mediate the cytotoxic effects of the β amyloid peptide Aβ 42 and serves as a receptor for humanin, a peptide that protects neuronal cells from damage by Aβ 42 , implying its involvement in the pathogenesis of Alzheimer's disease (AD). However, the interaction pattern between FPR2 and Aβ 42 or humanin remains unknown. Here we report the structures of FPR2 bound to G i and Aβ 42 or N-formyl humanin (fHN). Combined with functional data, the structures reveal two critical regions that govern recognition and activity of Aβ 42 and fHN, including a polar binding cavity within the receptor helical bundle and a hydrophobic binding groove in the extracellular region. In addition, the structures of FPR2 and FPR1 in complex with different formyl peptides were determined, providing insights into ligand recognition and selectivity of the FPR family. These findings uncover key factors that define the functionality of FPR2 in AD and other inflammatory diseases and would enable drug development.
Organizational Affiliation:
CAS Key Laboratory of Receptor Research, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.