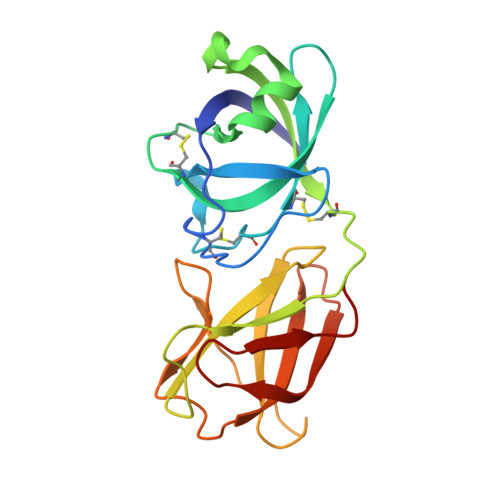
Boosting enzymatic degradation of cellulose using a fungal expansin: Structural insight into the pretreatment mechanism
Ding, S., Liu, X., Hakulinen, N., Taherzadeh, M.J., Wang, Y., Wang, Y., Qin, X., Wang, X., Yao, B., Luo, H., Tu, T.(2022) Bioresour Technol 358: 127434