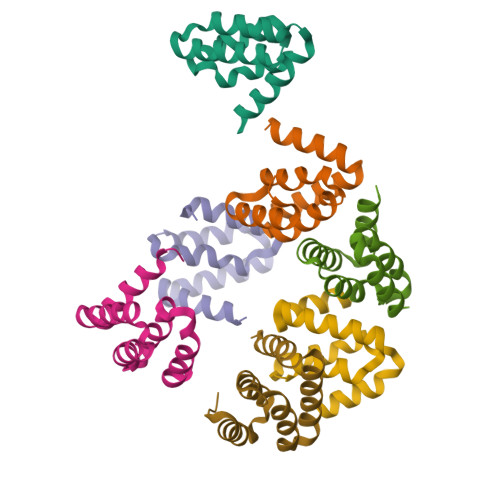
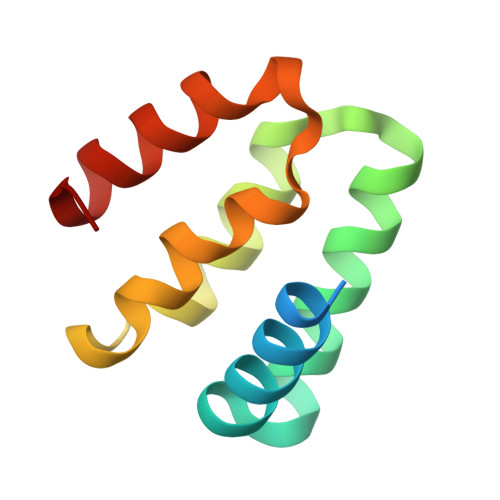
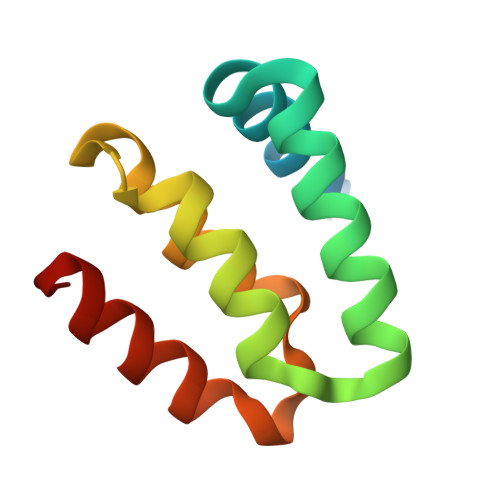
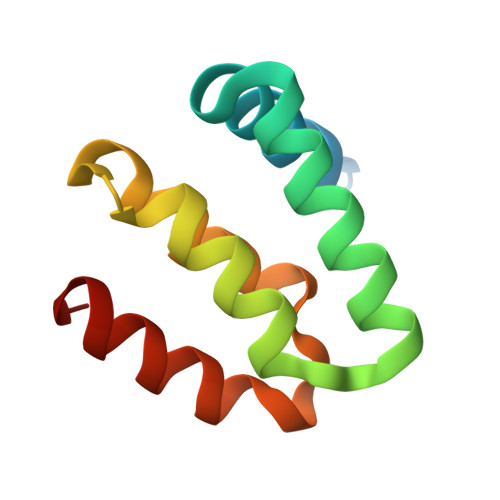
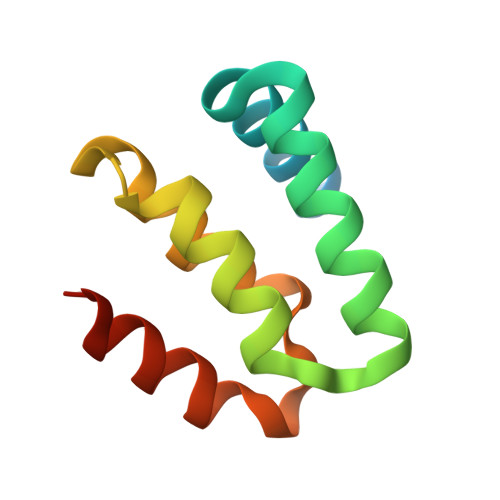
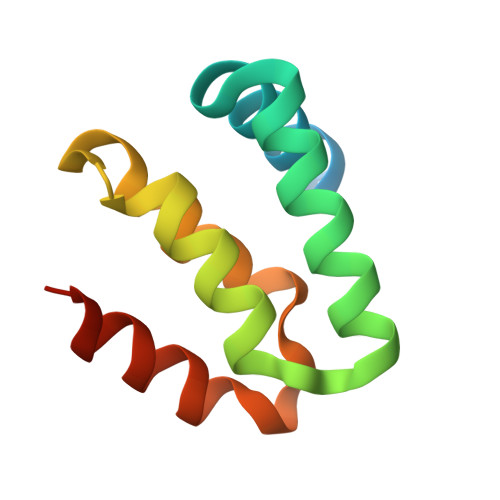
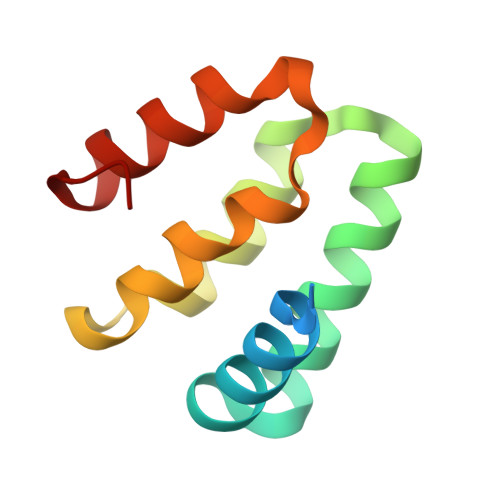
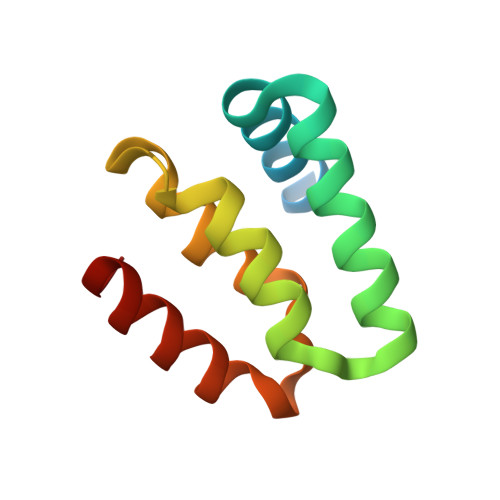
Harmonin homology domain-mediated interaction of RTEL1 helicase with RPA and DNA provides insights into its recruitment to DNA repair sites.
Kumar, N., Taneja, A., Ghosh, M., Rothweiler, U., Sundaresan, N.R., Singh, M.(2024) Nucleic Acids Res 52: 1450-1470
- PubMed: 38153196
- DOI: https://doi.org/10.1093/nar/gkad1208
- Primary Citation of Related Structures:
7WU8 - PubMed Abstract:
The regulator of telomere elongation helicase 1 (RTEL1) plays roles in telomere DNA maintenance, DNA repair, and genome stability by dismantling D-loops and unwinding G-quadruplex structures. RTEL1 comprises a helicase domain, two tandem harmonin homology domains 1&2 (HHD1 and HHD2), and a Zn2+-binding RING domain. In vitro D-loop disassembly by RTEL1 is enhanced in the presence of replication protein A (RPA). However, the mechanism of RTEL1 recruitment at non-telomeric D-loops remains unknown. In this study, we have unravelled a direct physical interaction between RTEL1 and RPA. Under DNA damage conditions, we showed that RTEL1 and RPA colocalise in the cell. Coimmunoprecipitation showed that RTEL1 and RPA interact, and the deletion of HHDs of RTEL1 significantly reduced this interaction. NMR chemical shift perturbations (CSPs) showed that RPA uses its 32C domain to interact with the HHD2 of RTEL1. Interestingly, HHD2 also interacted with DNA in the in vitro experiments. HHD2 structure was determined using X-ray crystallography, and NMR CSPs mapping revealed that both RPA 32C and DNA competitively bind to HHD2 on an overlapping surface. These results establish novel roles of accessory HHDs in RTEL1's functions and provide mechanistic insights into the RPA-mediated recruitment of RTEL1 to DNA repair sites.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bengaluru 560012, India.