Structural basis for assembly and disassembly of the IGF/IGFBP/ALS ternary complex
Kim, H., Fu, Y., Hong, H.J., Lee, S.G., Lee, D.S., Kim, H.M.(2022) Nat Commun 13: 4434
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Insulin-like growth factor-binding protein complex acid labile subunit | 578 | Homo sapiens | Mutation(s): 0 Gene Names: ALS | 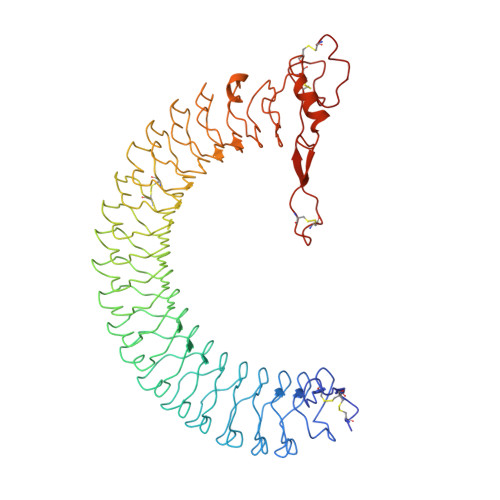 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P35858 (Homo sapiens) Explore P35858 Go to UniProtKB: P35858 | |||||
PHAROS: P35858 GTEx: ENSG00000099769 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35858 | ||||
Glycosylation | |||||
| Glycosylation Sites: 5 | Go to GlyGen: P35858-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Insulin-like growth factor-binding protein 3 | 264 | Homo sapiens | Mutation(s): 0 Gene Names: IGFBP3 | 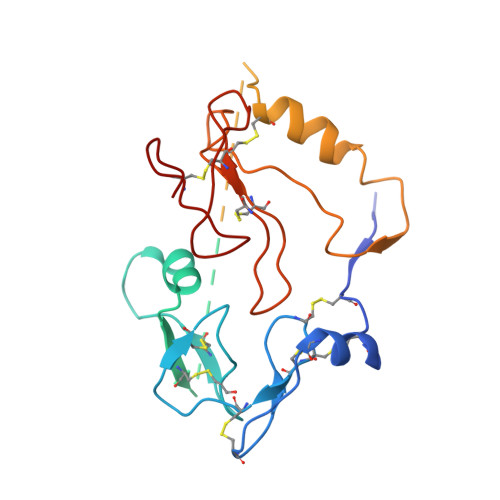 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P17936 (Homo sapiens) Explore P17936 Go to UniProtKB: P17936 | |||||
PHAROS: P17936 GTEx: ENSG00000146674 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P17936 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Isoform 3 of Insulin-like growth factor I | 70 | Homo sapiens | Mutation(s): 0 Gene Names: IGF1 | 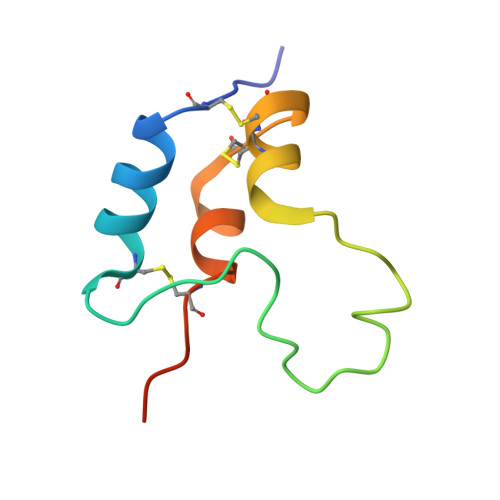 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P05019 (Homo sapiens) Explore P05019 Go to UniProtKB: P05019 | |||||
PHAROS: P05019 GTEx: ENSG00000017427 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05019 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | E [auth A], F [auth A], G [auth A], H [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | Institure of Basic Science, IBS-R030-C1 |