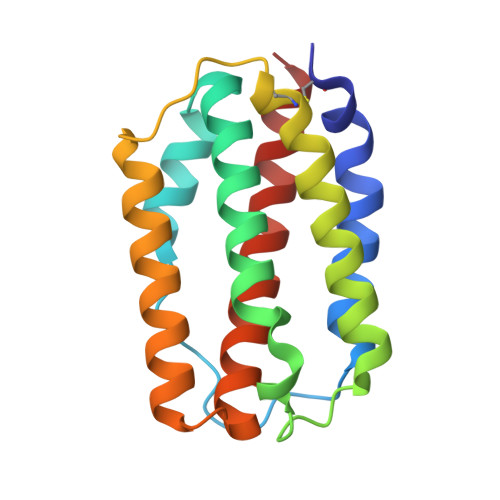
Structural and Functional Analyses of Type I IFNa Shed Light Into Its Interaction With Multiple Receptors in Fish.
Wang, Z., Xu, J., Feng, J., Wu, K., Chen, K., Jia, Z., Zhu, X., Huang, W., Zhao, X., Liu, Q., Wang, B., Chen, X., Wang, J., Zou, J.(2022) Front Immunol 13: 862764-862764
- PubMed: 35392096
- DOI: https://doi.org/10.3389/fimmu.2022.862764
- Primary Citation of Related Structures:
7WKH - PubMed Abstract:
Teleost type I interferons (IFNs) are categorized into group I and II subgroups that bind to distinct receptors to activate antiviral responses. However, the interaction between ifn ligands and receptors has not fully been understood. In this study, the crystal structure of grass carp [ Ctenopharyngodon idella ( Ci )] IFNa has been solved at 1.58Å and consists of six helices. The Ci IFNa displays a typical structure of type I IFNs with a straight helix F and lacks a helix element in the AB loop. Superposition modeling identified several key residues involved in the interaction with receptors. It was found that Ci IFNa bound to cytokine receptor family B (CRFB) 1, CRFB2, and CRFB5, and the three receptors could form heterodimeric receptor complexes. Furthermore, mutation of Leu27, Glu103, Lys117, and His165 markedly decreased the phosphorylation of signal transducer and activator of transcription (STAT) 1a induced by Ci IFNa in the Epithelioma papulosum cyprini (EPC) cells, and Glu103 was shown to be required for the Ci IFNa-activated antiviral activity. Interestingly, wild-type and mutant Ci IFNa proteins did not alter the phosphorylation levels of STAT1b. Our results demonstrate that fish type I IFNs, although structurally conserved, interact with the receptors in a manner that may differ from mammalian homologs.
Organizational Affiliation:
Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, China.