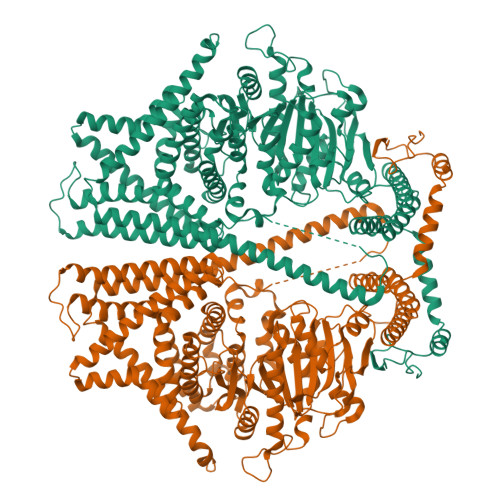
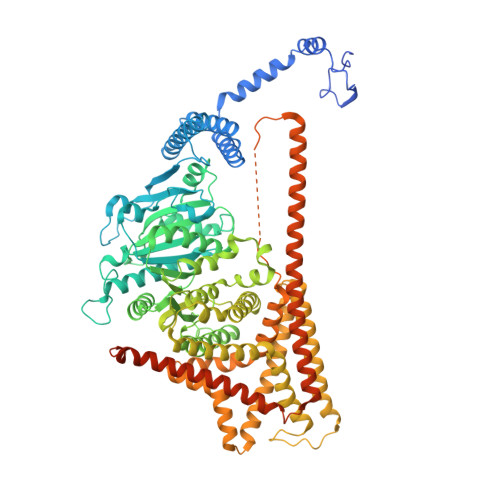
Structural basis for directional chitin biosynthesis.
Chen, W., Cao, P., Liu, Y., Yu, A., Wang, D., Chen, L., Sundarraj, R., Yuchi, Z., Gong, Y., Merzendorfer, H., Yang, Q.(2022) Nature 610: 402-408
- PubMed: 36131020
- DOI: https://doi.org/10.1038/s41586-022-05244-5
- Primary Citation of Related Structures:
7WJM, 7WJN, 7WJO, 7X05, 7X06 - PubMed Abstract:
Chitin, the most abundant aminopolysaccharide in nature, is an extracellular polymer consisting of N-acetylglucosamine (GlcNAc) units 1 . The key reactions of chitin biosynthesis are catalysed by chitin synthase 2-4 , a membrane-integrated glycosyltransferase that transfers GlcNAc from UDP-GlcNAc to a growing chitin chain. However, the precise mechanism of this process has yet to be elucidated. Here we report five cryo-electron microscopy structures of a chitin synthase from the devastating soybean root rot pathogenic oomycete Phytophthora sojae (PsChs1). They represent the apo, GlcNAc-bound, nascent chitin oligomer-bound, UDP-bound (post-synthesis) and chitin synthase inhibitor nikkomycin Z-bound states of the enzyme, providing detailed views into the multiple steps of chitin biosynthesis and its competitive inhibition. The structures reveal the chitin synthesis reaction chamber that has the substrate-binding site, the catalytic centre and the entrance to the polymer-translocating channel that allows the product polymer to be discharged. This arrangement reflects consecutive key events in chitin biosynthesis from UDP-GlcNAc binding and polymer elongation to the release of the product. We identified a swinging loop within the chitin-translocating channel, which acts as a 'gate lock' that prevents the substrate from leaving while directing the product polymer into the translocating channel for discharge to the extracellular side of the cell membrane. This work reveals the directional multistep mechanism of chitin biosynthesis and provides a structural basis for inhibition of chitin synthesis.
Organizational Affiliation:
State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China.