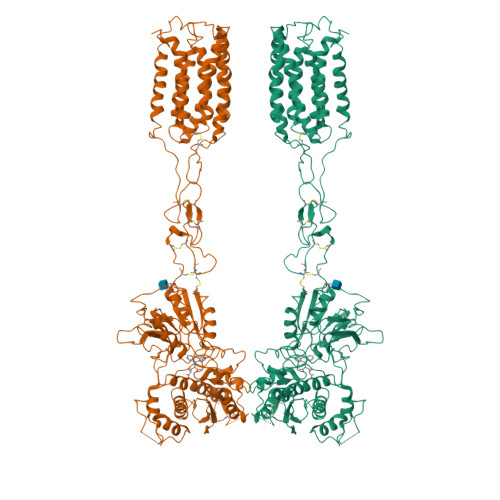
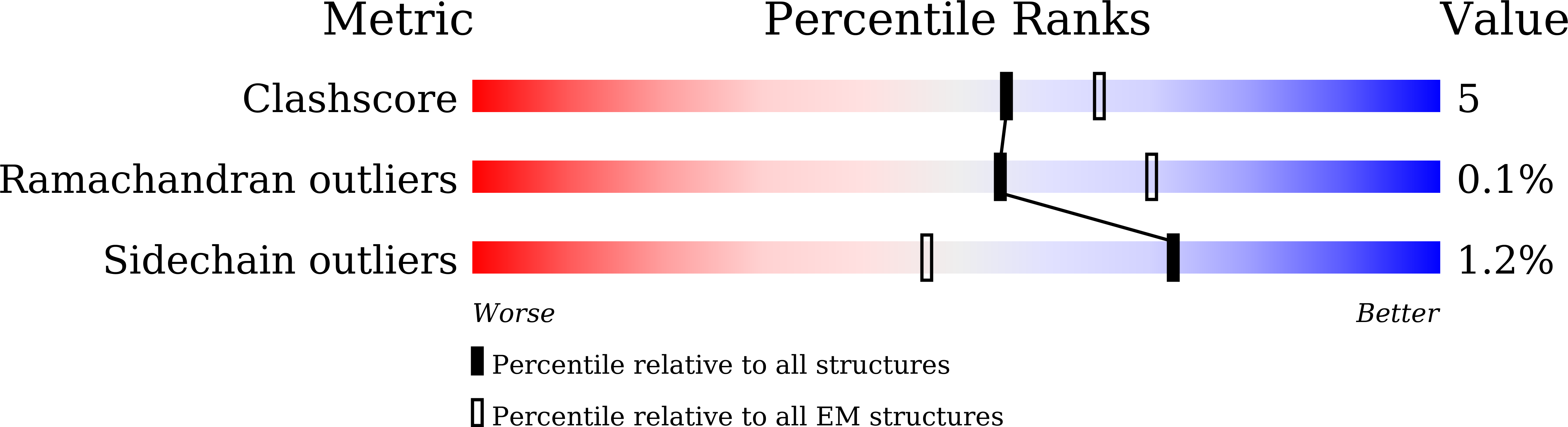Structural basis of the activation of metabotropic glutamate receptor 3.
Fang, W., Yang, F., Xu, C., Ling, S., Lin, L., Zhou, Y., Sun, W., Wang, X., Liu, P., Rondard, P., Shi, P., Pin, J.P., Tian, C., Liu, J.(2022) Cell Res 32: 695-698
- PubMed: 35236939
- DOI: https://doi.org/10.1038/s41422-022-00623-z
- Primary Citation of Related Structures:
7WI6, 7WI8, 7WIH
Organizational Affiliation:
The First Affiliated Hospital of USTC, School of Life Sciences, Division of Life Sciences and Medicine, Joint Center for Biological Analytical Chemistry, Anhui Engineering Laboratory of Peptide Drug, Anhui Laboratory of Advanced Photonic Science and Technology, University of Science and Technology of China, Hefei, Anhui, China.