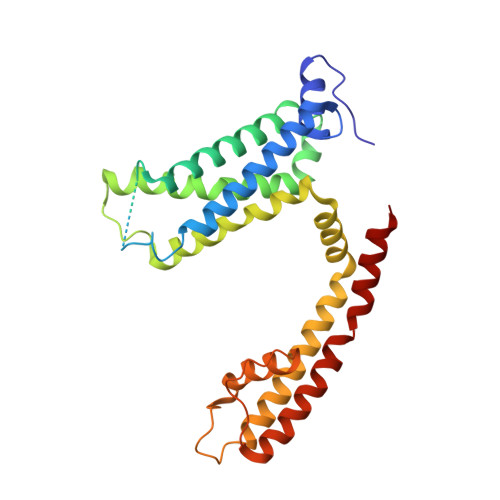
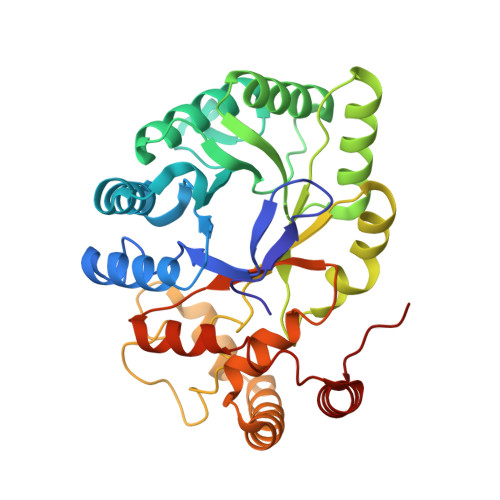
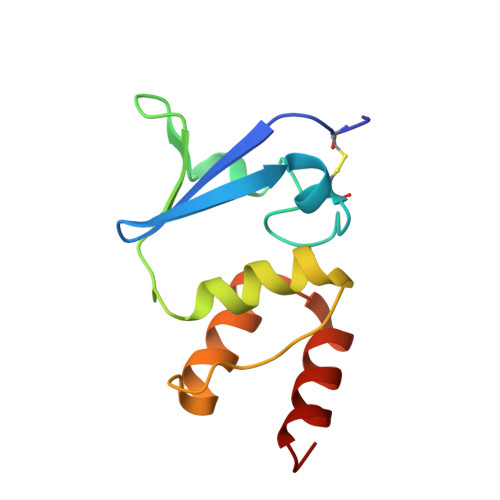
Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Tyagi, A., Ahmed, T., Jian, S., Bajaj, S., Ong, S.T., Goay, S.S.M., Zhao, Y., Vorobyov, I., Tian, C., Chandy, K.G., Bhushan, S.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35091471
- DOI: https://doi.org/10.1073/pnas.2113536119
- Primary Citation of Related Structures:
7WF3, 7WF4 - PubMed Abstract:
We report two structures of the human voltage-gated potassium channel (Kv) Kv1.3 in immune cells alone (apo-Kv1.3) and bound to an immunomodulatory drug called dalazatide (dalazatide-Kv1.3). Both the apo-Kv1.3 and dalazatide-Kv1.3 structures are in an activated state based on their depolarized voltage sensor and open inner gate. In apo-Kv1.3, the aromatic residue in the signature sequence (Y447) adopts a position that diverges 11 Å from other K + channels. The outer pore is significantly rearranged, causing widening of the selectivity filter and perturbation of ion binding within the filter. This conformation is stabilized by a network of intrasubunit hydrogen bonds. In dalazatide-Kv1.3, binding of dalazatide to the channel's outer vestibule narrows the selectivity filter, Y447 occupies a position seen in other K + channels, and this conformation is stabilized by a network of intersubunit hydrogen bonds. These remarkable rearrangements in the selectivity filter underlie Kv1.3's transition into the drug-blocked state.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, Singapore 637551.