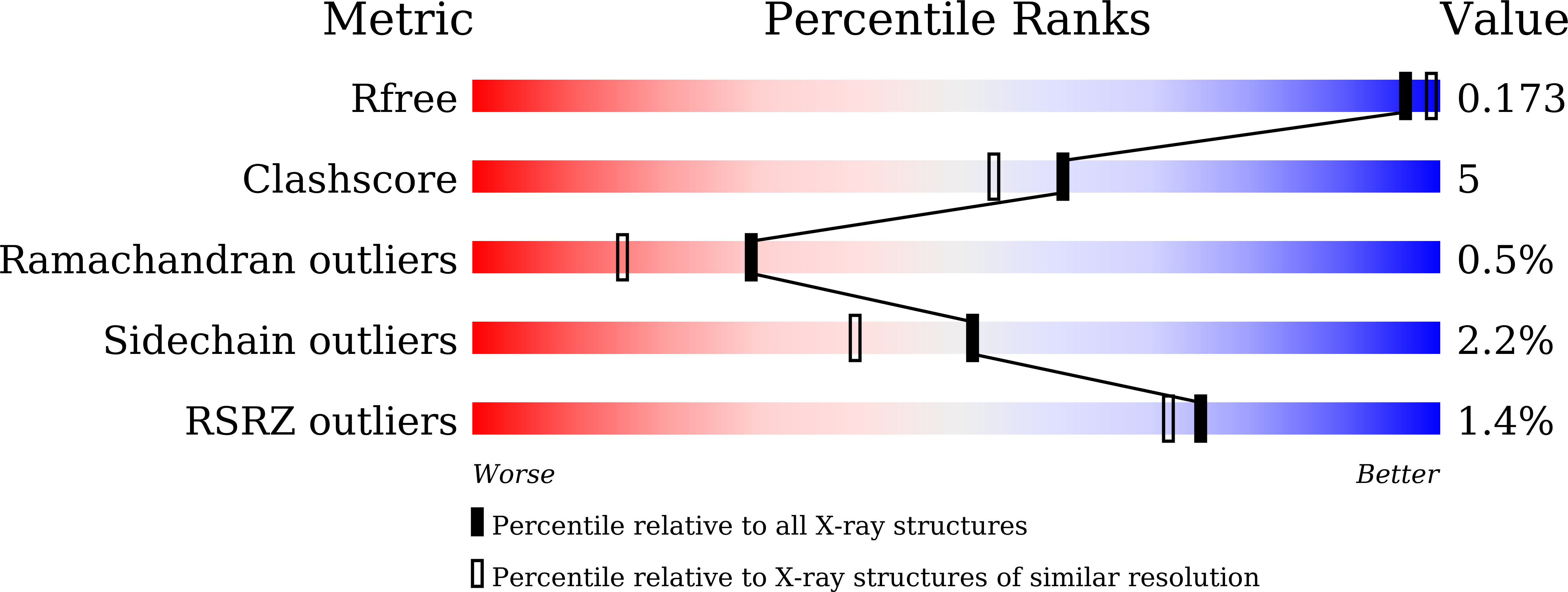
Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides.
Matsuzawa, T., Watanabe, M., Nakamichi, Y., Akita, H., Yaoi, K.(2022) Appl Microbiol Biotechnol 106: 4539-4551
- PubMed: 35723691
- DOI: https://doi.org/10.1007/s00253-022-12018-6
- Primary Citation of Related Structures:
7WDN - PubMed Abstract:
Metagenomic MeBglD2 is a glycoside hydrolase family 1 (GH1) β-glycosidase that has β-glucosidase, β-fucosidase, and β-galactosidase activities, and is highly activated in the presence of monosaccharides and disaccharides. The β-glucosidase activity of MeBglD2 increases in a cellobiose concentration-dependent manner and is not inhibited by a high concentration of D-glucose or cellobiose. Previously, we solved the crystal structure of MeBglD2 and designed a thermostable mutant; however, the mechanism of substrate recognition of MeBglD2 remains poorly understood. In this paper, we report the X-ray crystal structures of MeBglD2 complexed with various saccharides, such as D-glucose, D-xylose, cellobiose, and maltose. The results showed that subsite - 1 of MeBglD2, which contained two catalytic glutamate residues (a nucleophilic Glu356 and an acid/base Glu170) was common to other GH1 enzymes, but the positive subsites (+ 1 and + 2) had different binding modes depending on the type of sugar. Three residues (Glu183, Asn227, and Asn229), located at the positive subsites of MeBglD2, were involved in substrate specificity toward cellobiose and/or chromogenic substrates in the presence of additive sugars. The docking simulation of MeBglD2-cellobiose indicated that Asn229 and Trp329 play important roles in the recognition of + 1 D-glucose in cellobiose. Our findings provide insights into the unique substrate recognition mechanism of GH1, which can incorporate a variety of saccharides into its positive subsites. KEY POINTS: • Metagenomic glycosidase, MeBglD2, recognizes various saccharides • Structures of metagenomic MeBglD2 complexed with various saccharides are determined • MeBglD2 has a unique substrate recognition mechanism at the positive subsites.
Organizational Affiliation:
Department of Applied Biological Science, Faculty of Agriculture, Kagawa University, 2393 Ikenobe, Miki-cho, Kita-gun, Kagawa, 761-0795, Japan. matsuzawa.tomohiko@kagawa-u.ac.jp.