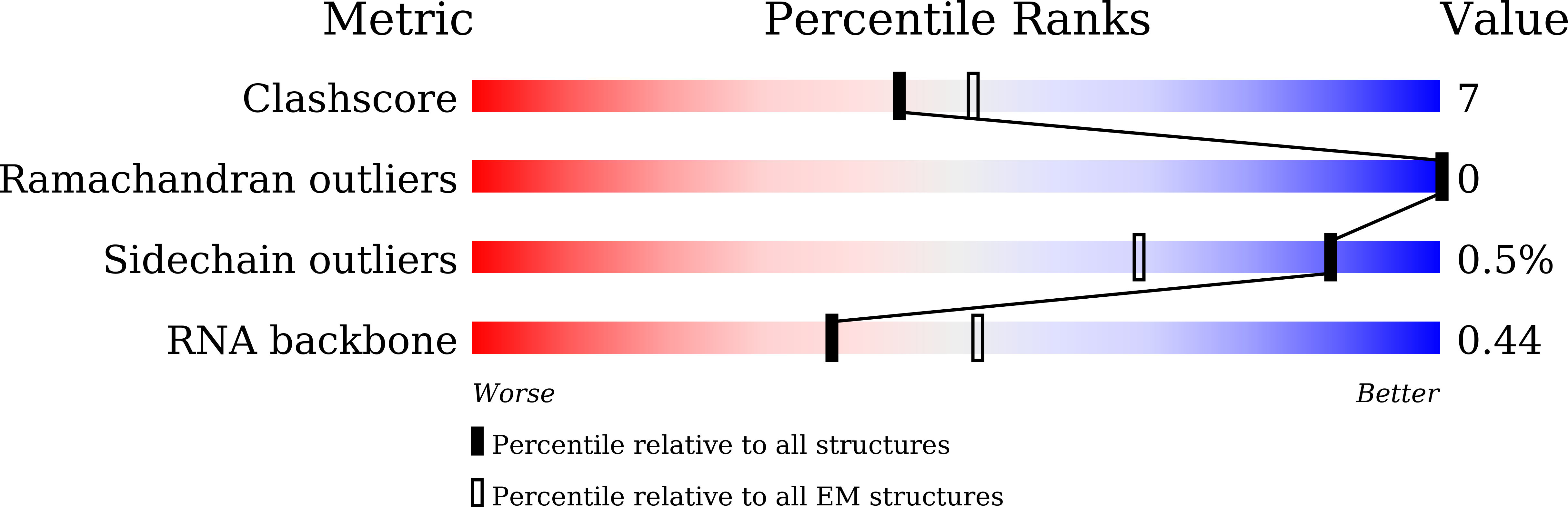
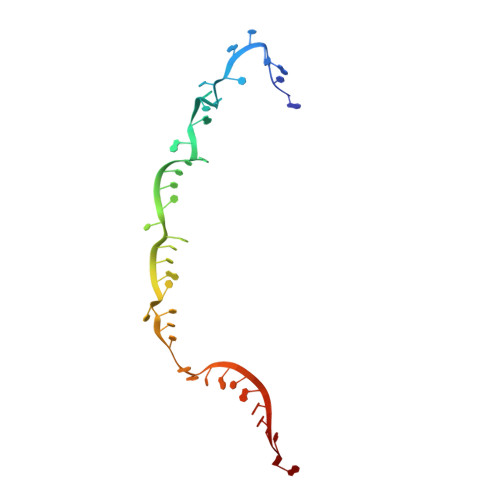
Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex.
Kato, K., Zhou, W., Okazaki, S., Isayama, Y., Nishizawa, T., Gootenberg, J.S., Abudayyeh, O.O., Nishimasu, H.(2022) Cell 185: 2324-2337.e16
- PubMed: 35643083
- DOI: https://doi.org/10.1016/j.cell.2022.05.003
- Primary Citation of Related Structures:
7WAH - PubMed Abstract:
The type III-E CRISPR-Cas effector Cas7-11, with dual RNase activities for precursor CRISPR RNA (pre-crRNA) processing and crRNA-guided target RNA cleavage, is a new platform for bacterial and mammalian RNA targeting. We report the 2.5-Å resolution cryoelectron microscopy structure of Cas7-11 in complex with a crRNA and its target RNA. Cas7-11 adopts a modular architecture comprising seven domains (Cas7.1-Cas7.4, Cas11, INS, and CTE) and four interdomain linkers. The crRNA 5' tag is recognized and processed by Cas7.1, whereas the crRNA spacer hybridizes with the target RNA. Consistent with our biochemical data, the catalytic residues for programmable cleavage in Cas7.2 and Cas7.3 neighbor the scissile phosphates before the flipped-out fourth and tenth nucleotides in the target RNA, respectively. Using structural insights, we rationally engineered a compact Cas7-11 variant (Cas7-11S) for single-vector AAV packaging for transcript knockdown in human cells, enabling in vivo Cas7-11 applications.
Organizational Affiliation:
Structural Biology Division, Research Center for Advanced Science and Technology, The University of Tokyo, 4-6-1 Komaba, Meguro-ku, Tokyo 153-8904, Japan.