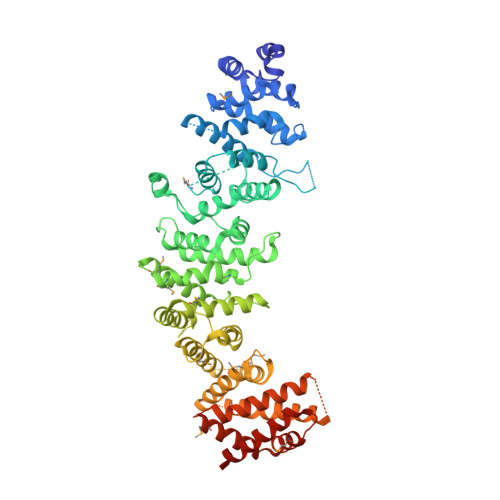
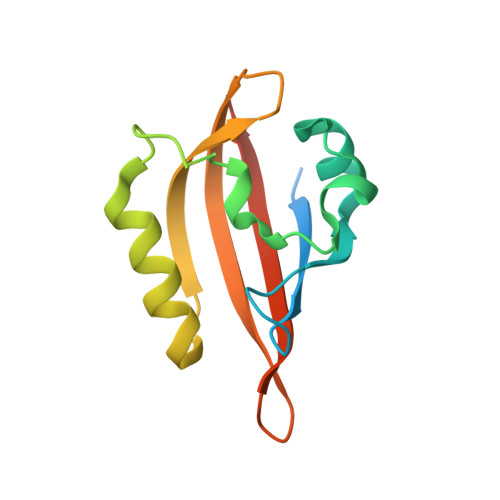
Structural analysis of the regulation of blue-light receptors by GIGANTEA.
Kwon, E., Pathak, D., Dahal, P., Tandukar, S., Jung, H.S., Kim, W.Y., Kim, D.Y.(2022) Cell Rep 39: 110700-110700
- PubMed: 35443175
- DOI: https://doi.org/10.1016/j.celrep.2022.110700
- Primary Citation of Related Structures:
7WA4 - PubMed Abstract:
In Arabidopsis, GIGANTEA (GI), together with the blue-light receptors ZTL, LKP2, and FKF1, regulates degradation of the core clock protein TOC1 and the flowering repressor CDFs, thereby controlling circadian oscillation and flowering. Despite the significance of GI in diverse plant physiology, its molecular function is not much understood because of technical problems in protein preparation and a lack of structural information. Here, we report the purification of the GI monomer and the crystal structure of the GI/LKP2 complex. The crystal structure reveals that residues 1-813 of GI possess an elongated rigid structure formed by stacking hydrophobic α-helices and that the LOV domain of LKP2 binds to the middle region of the GI (residues 563-789). Interaction analysis further shows that LOV homodimers are converted to monomers by GI binding. Our results provide structural insights into the regulation of the circadian clock and photoperiodic flowering by GI and ZTL/LKP2/FKF1.
Organizational Affiliation:
College of Pharmacy, Yeungnam University, Gyeongsan 38541, Republic of Korea.