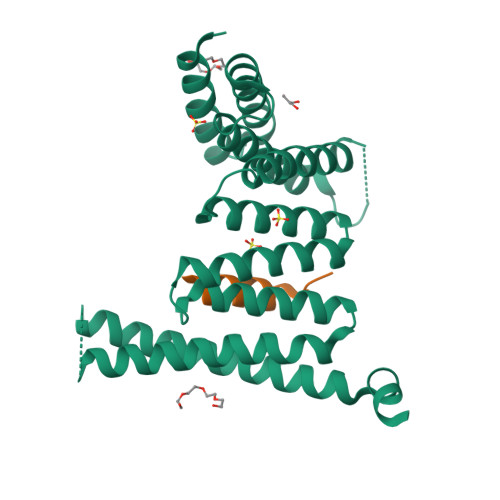
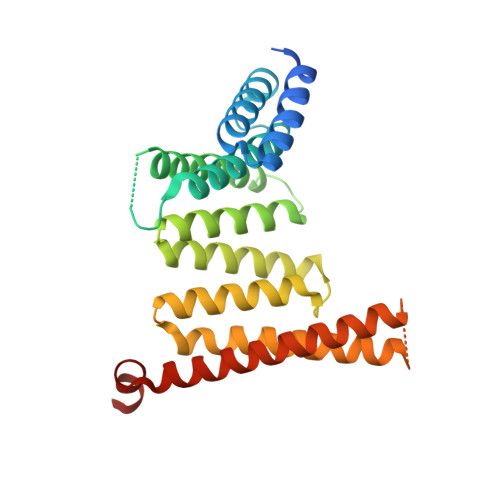
Structural basis for histone H3 recognition by NASP in Arabidopsis.
Liu, Y., Chen, L., Wang, N., Wu, B., Bao, H., Huang, H.(2022) J Integr Plant Biol 64: 2309-2313
- PubMed: 35587028
- DOI: https://doi.org/10.1111/jipb.13277
- Primary Citation of Related Structures:
7W5M - PubMed Abstract:
The structural basis for histone recognition by the histone chaperone nuclear autoantigenic sperm protein (NASP) remains largely unclear. Here, we showed that Arabidopsis thaliana AtNASP is a monomer and displays robust nucleosome assembly activity in vitro. Examining the structure of AtNASP complexed with a histone H3 α3 peptide revealed a binding mode that is conserved in human NASP. AtNASP recognizes the H3 N-terminal region distinct from human NASP. Moreover, AtNASP forms a co-chaperone complex with ANTI-SILENCING FUNCTION 1 (ASF1) by binding to the H3 N-terminal region. Therefore, we deciphered the structure of AtNASP and the basis of the AtNASP-H3 interaction.
Organizational Affiliation:
Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes, Department of Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen, 518055, China.