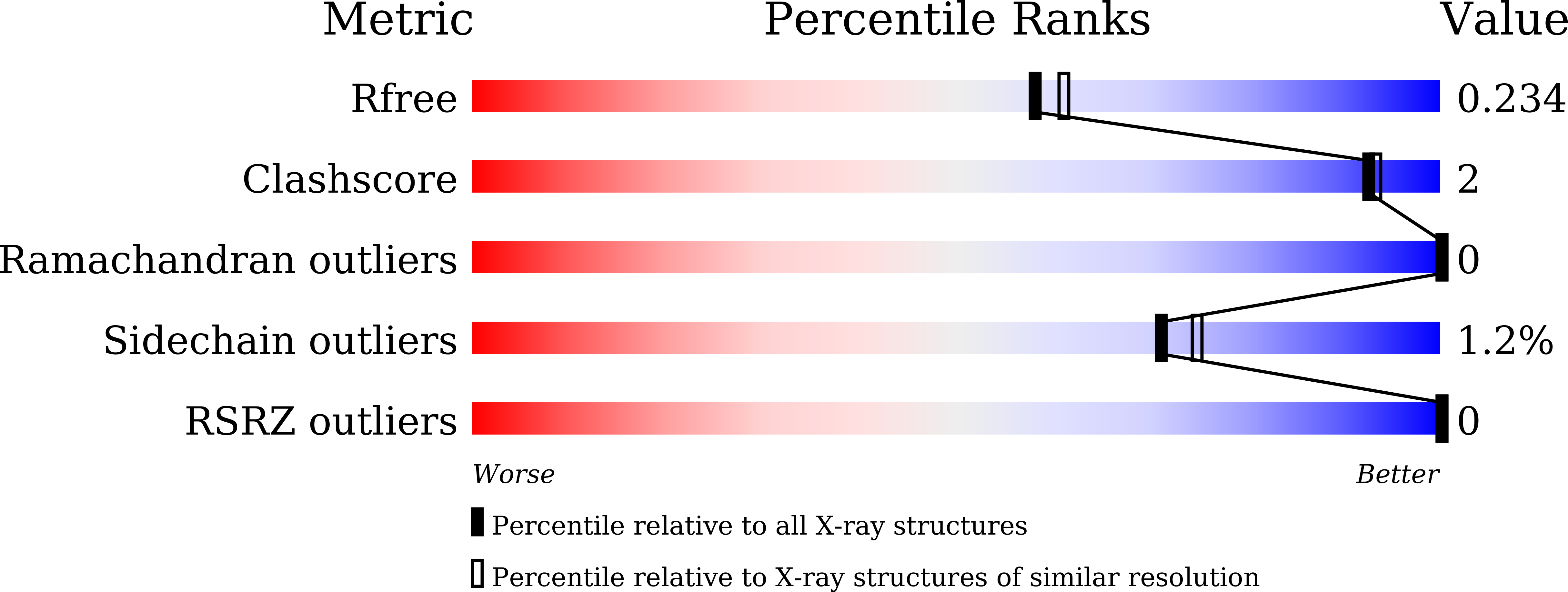
Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
Xu, L., Wang, J., Xiao, Y., Han, Z., Chai, J.(2023) J Integr Plant Biol 65: 235-248
- PubMed: 35568972
- DOI: https://doi.org/10.1111/jipb.13279
- Primary Citation of Related Structures:
7VS7 - PubMed Abstract:
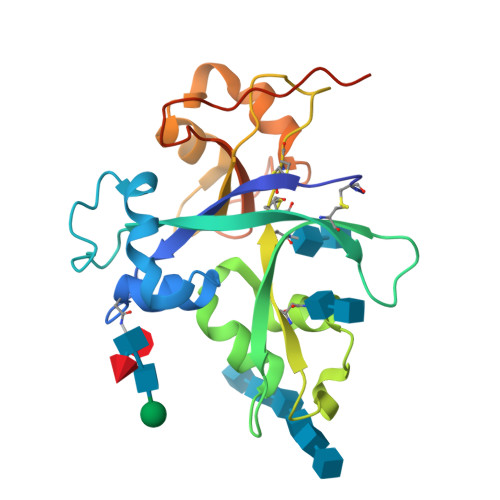
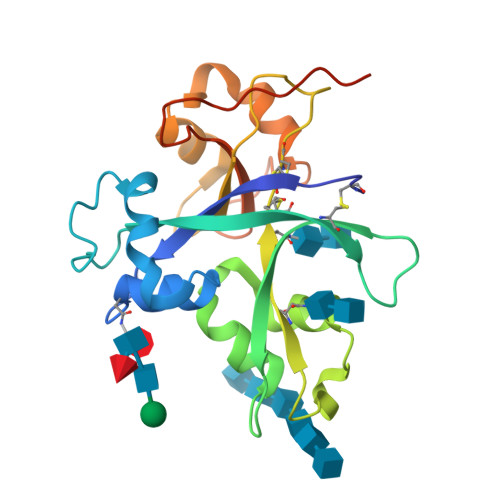
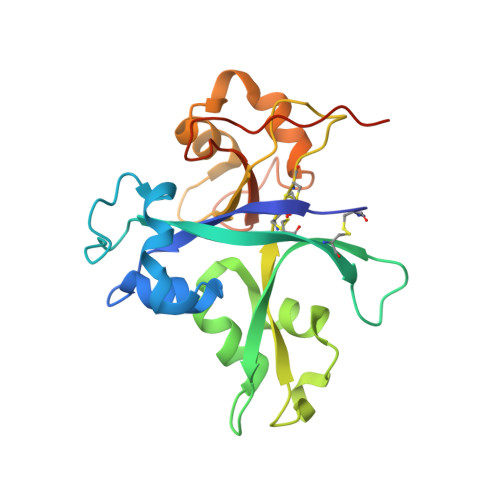
Plants have developed innate immune systems to fight against pathogenic fungi by monitoring pathogenic signals known as pathogen-associated molecular patterns (PAMP) and have established endo symbiosis with arbuscular mycorrhizal (AM) fungi through recognition of mycorrhizal (Myc) factors. Chitin elicitor receptor kinase 1 of Oryza sativa subsp. Japonica (OsCERK1) plays a bifunctional role in mediating both chitin-triggered immunity and symbiotic relationships with AM fungi. However, it remains unclear whether OsCERK1 can directly recognize chitin molecules. In this study, we show that OsCERK1 binds to the chitin hexamer ((NAG) 6 ) and tetramer ((NAG) 4 ) directly and determine the crystal structure of the OsCERK1-(NAG) 6 complex at 2 Å. The structure shows that one OsCERK1 is associated with one (NAG) 6 . Upon recognition, chitin hexamer binds OsCERK1 by interacting with the shallow groove on the surface of LysM2. These structural findings, complemented by mutational analyses, demonstrate that LysM2 is crucial for recognition of both (NAG) 6 and (NAG) 4 . Altogether, these findings provide structural insights into the ability of OsCERK1 in chitin perception, which will lead to a better understanding of the role of OsCERK1 in mediating both immunity and symbiosis in rice.
Organizational Affiliation:
Tsinghua-Peking Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, Centre for Plant Biology, School of Life Sciences, Tsinghua University, Beijing, 100084, China.