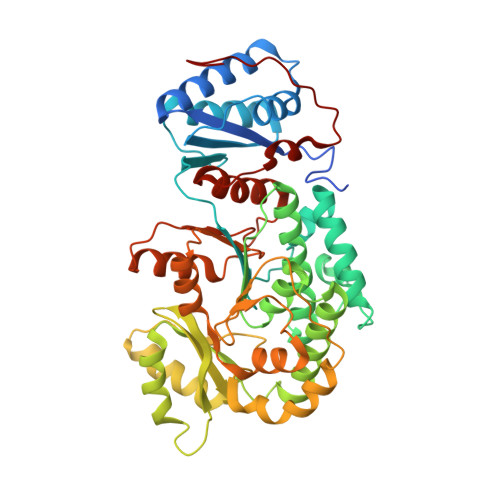
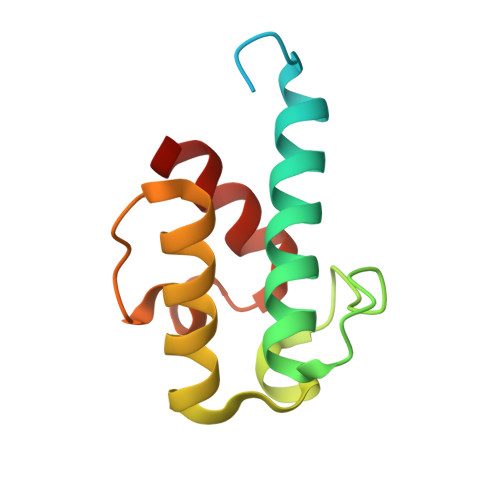
Structural visualization of transient interactions between the cis-acting acyltransferase and acyl carrier protein of the salinomycin modular polyketide synthase.
Feng, Y., Zhang, F., Huang, S., Deng, Z., Bai, L., Zheng, J.(2022) Acta Crystallogr D Struct Biol 78: 779-791
- PubMed: 35647924
- DOI: https://doi.org/10.1107/S2059798322004612
- Primary Citation of Related Structures:
7VRS, 7VT1 - PubMed Abstract:
Transient protein-protein interactions between cis-acting acyltransferase (AT) and acyl carrier protein (ACP) domains are critical for the catalysis and processivity of modular polyketide synthases (mPKSs), but are challenging for structural characterization due to the intrinsically weak binding affinity. Here, a stable complex of cis-acting AT and ACP domains from the ninth module of the salinomycin mPKS was obtained using a maleimide cross-linker and the structure of the complex was determined at 2.6 Å resolution. The crystal structure shows that the AT in combination with the ketosynthase (KS)-to-AT linker forms a C-shaped architecture to embrace the ACP. The large hydrolase subdomain of the AT serves as a major binding platform for the ACP, while the small ferredoxin-like subdomain of the AT and the KS-to-AT linker cooperate with each other to constrain binding of the ACP. The importance of interface residues in cis-acting AT-ACP interactions was confirmed by mutagenesis assays. The interaction mode observed in the cis-acting AT-ACP complex is completely different from those observed in trans-acting AT-ACP complexes, where the ACP primarily contacts the small domain of the AT. The complex structure provides detailed mechanistic insights into AT-ACP recognition in cis-AT mPKSs.
Organizational Affiliation:
State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, 800 Dongchuan Road, Minhang District, Shanghai 200240, People's Republic of China.