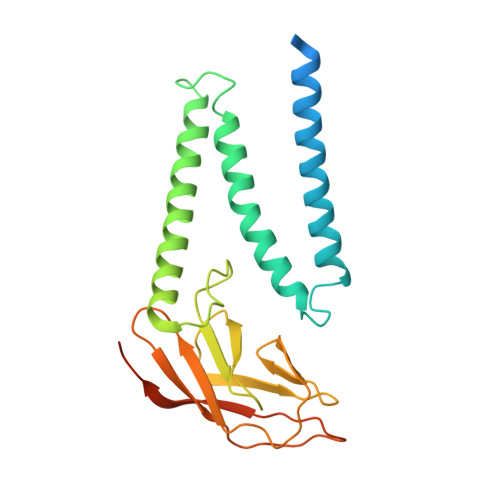
Structure of SARS-CoV-2 membrane protein essential for virus assembly.
Zhang, Z., Nomura, N., Muramoto, Y., Ekimoto, T., Uemura, T., Liu, K., Yui, M., Kono, N., Aoki, J., Ikeguchi, M., Noda, T., Iwata, S., Ohto, U., Shimizu, T.(2022) Nat Commun 13: 4399-4399
- PubMed: 35931673
- DOI: https://doi.org/10.1038/s41467-022-32019-3
- Primary Citation of Related Structures:
7VGR, 7VGS - PubMed Abstract:
The coronavirus membrane protein (M) is the most abundant viral structural protein and plays a central role in virus assembly and morphogenesis. However, the process of M protein-driven virus assembly are largely unknown. Here, we report the cryo-electron microscopy structure of the SARS-CoV-2 M protein in two different conformations. M protein forms a mushroom-shaped dimer, composed of two transmembrane domain-swapped three-helix bundles and two intravirion domains. M protein further assembles into higher-order oligomers. A highly conserved hinge region is key for conformational changes. The M protein dimer is unexpectedly similar to SARS-CoV-2 ORF3a, a viral ion channel. Moreover, the interaction analyses of M protein with nucleocapsid protein (N) and RNA suggest that the M protein mediates the concerted recruitment of these components through the positively charged intravirion domain. Our data shed light on the M protein-driven virus assembly mechanism and provide a structural basis for therapeutic intervention targeting M protein.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, Japan.