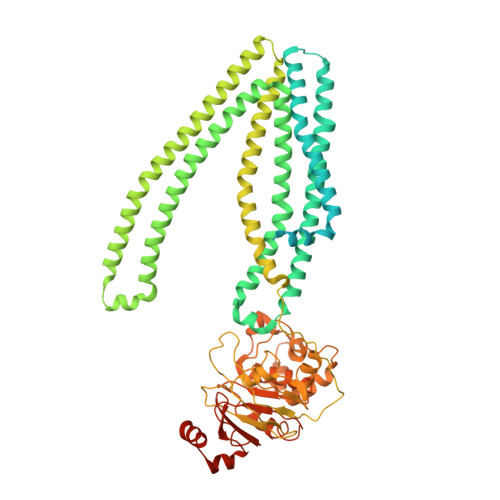
The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Park, J.G., Kim, S., Jang, E., Choi, S.H., Han, H., Ju, S., Kim, J.W., Min, D.S., Jin, M.S.(2022) Nat Commun 13: 5851-5851
- PubMed: 36195619
- DOI: https://doi.org/10.1038/s41467-022-33593-2
- Primary Citation of Related Structures:
7V5C, 7V5D, 7VFI - PubMed Abstract:
TAPL is a lysosomal ATP-binding cassette transporter that translocates a broad spectrum of polypeptides from the cytoplasm into the lysosomal lumen. Here we report that, in addition to its well-known role as a peptide translocator, TAPL exhibits an ATP-dependent phosphatidylserine floppase activity that is the possible cause of its high basal ATPase activity and of the lack of coupling between ATP hydrolysis and peptide efflux. We also present the cryo-EM structures of mouse TAPL complexed with (i) phospholipid, (ii) cholesteryl hemisuccinate (CHS) and 9-mer peptide, and (iii) ADP·BeF 3 . The inward-facing structure reveals that F449 protrudes into the cylindrical transport pathway and divides it into a large hydrophilic central cavity and a sizable hydrophobic upper cavity. In the structure, the peptide binds to TAPL in horizontally-stretched fashion within the central cavity, while lipid molecules plug vertically into the upper cavity. Together, our results suggest that TAPL uses different mechanisms to function as a peptide translocase and a phosphatidylserine floppase.
Organizational Affiliation:
School of Life Sciences, Gwangju Institute of Science and Technology (GIST), Gwangju, 61005, Republic of Korea.