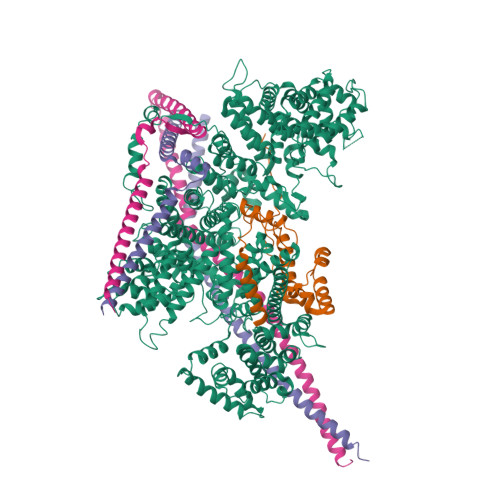
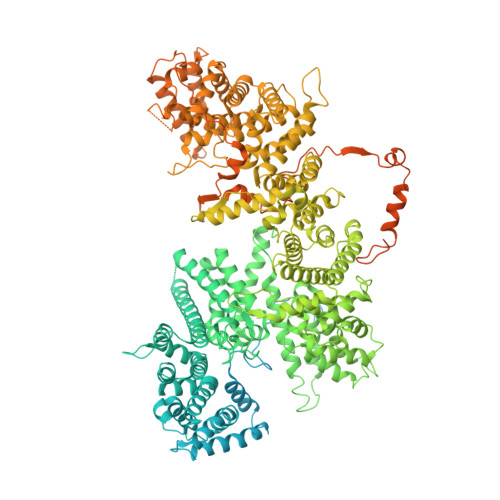
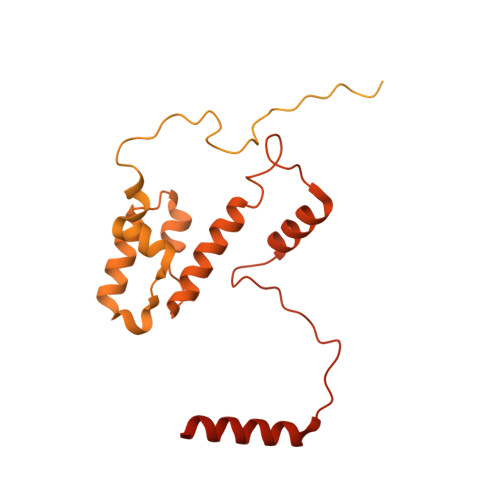
Cryo-EM structures of human m6A writer complexes.
Su, S., Li, S., Deng, T., Gao, M., Yin, Y., Wu, B., Peng, C., Liu, J., Ma, J., Zhang, K.(2022) Cell Res 32: 982-994
- PubMed: 36167981
- DOI: https://doi.org/10.1038/s41422-022-00725-8
- Primary Citation of Related Structures:
7VF2, 7VF5 - PubMed Abstract:
N 6 -methyladenosine (m 6 A) is the most abundant ribonucleotide modification among eukaryotic messenger RNAs. The m 6 A "writer" consists of the catalytic subunit m 6 A-METTL complex (MAC) and the regulatory subunit m 6 A-METTL-associated complex (MACOM), the latter being essential for enzymatic activity. Here, we report the cryo-electron microscopy (cryo-EM) structures of MACOM at a 3.0-Å resolution, uncovering that WTAP and VIRMA form the core structure of MACOM and that ZC3H13 stretches the conformation by binding VIRMA. Furthermore, the 4.4-Å resolution cryo-EM map of the MACOM-MAC complex, combined with crosslinking mass spectrometry and GST pull-down analysis, elucidates a plausible model of the m 6 A writer complex, in which MACOM binds to MAC mainly through WTAP and METTL3 interactions. In combination with in vitro RNA substrate binding and m 6 A methyltransferase activity assays, our results illustrate the molecular basis of how MACOM assembles and interacts with MAC to form an active m 6 A writer complex.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Multiscale Research Institute of Complex Systems, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai, China.