Structure, function and pharmacology of human itch receptor complexes.
Yang, F., Guo, L., Li, Y., Wang, G., Wang, J., Zhang, C., Fang, G.X., Chen, X., Liu, L., Yan, X., Liu, Q., Qu, C., Xu, Y., Xiao, P., Zhu, Z., Li, Z., Zhou, J., Yu, X., Gao, N., Sun, J.P.(2021) Nature 600: 164-169
- PubMed: 34789875
- DOI: https://doi.org/10.1038/s41586-021-04077-y
- Primary Citation of Related Structures:
7VDH, 7VDL, 7VDM, 7VUY, 7VUZ, 7VV0, 7VV3, 7VV4, 7VV5, 7VV6 - PubMed Abstract:
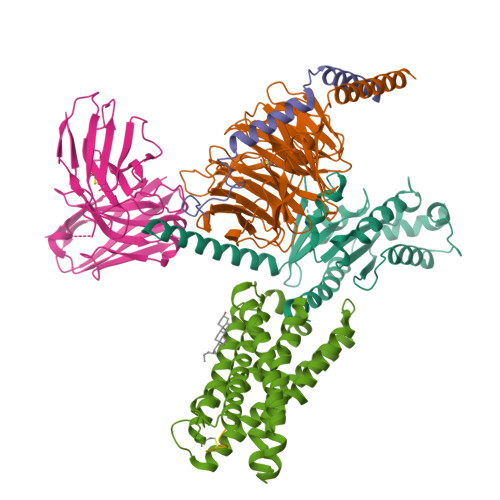
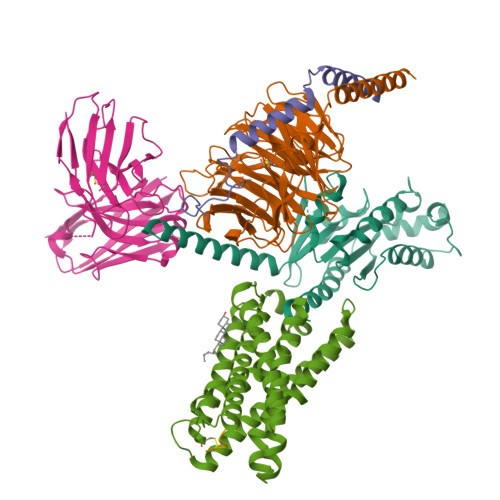
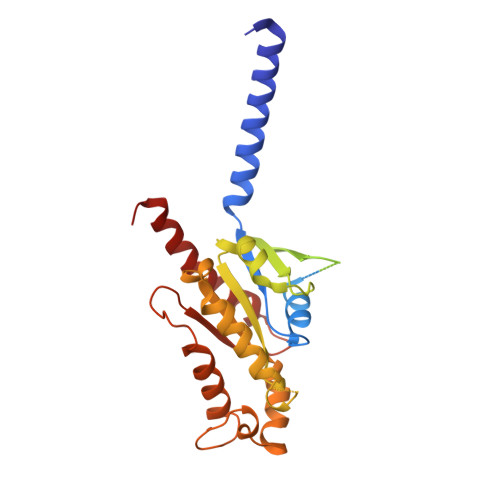
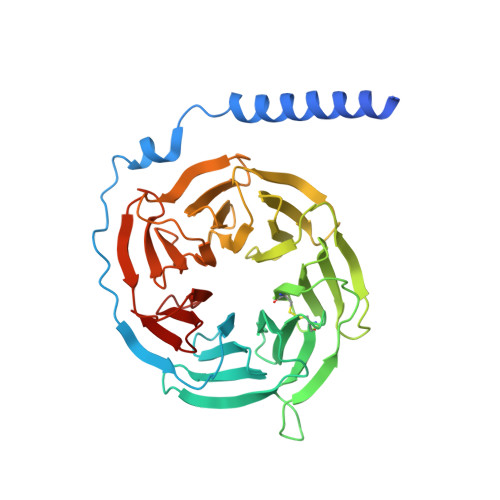
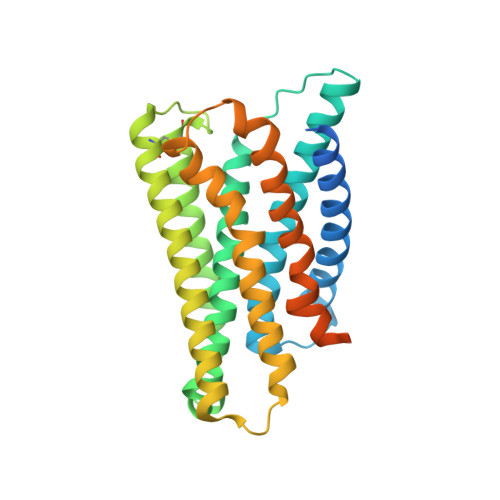
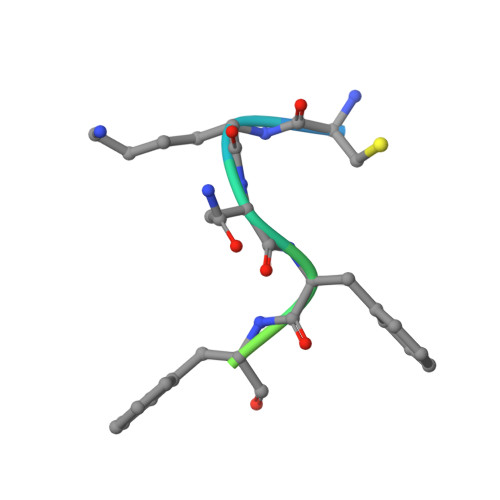
In the clades of animals that diverged from the bony fish, a group of Mas-related G-protein-coupled receptors (MRGPRs) evolved that have an active role in itch and allergic signals 1,2 . As an MRGPR, MRGPRX2 is known to sense basic secretagogues (agents that promote secretion) and is involved in itch signals and eliciting pseudoallergic reactions 3-6 . MRGPRX2 has been targeted by drug development efforts to prevent the side effects induced by certain drugs or to treat allergic diseases. Here we report a set of cryo-electron microscopy structures of the MRGPRX2-G i1 trimer in complex with polycationic compound 48/80 or with inflammatory peptides. The structures of the MRGPRX2-G i1 complex exhibited shallow, solvent-exposed ligand-binding pockets. We identified key common structural features of MRGPRX2 and describe a consensus motif for peptidic allergens. Beneath the ligand-binding pocket, the unusual kink formation at transmembrane domain 6 (TM6) and the replacement of the general toggle switch from Trp 6.48 to Gly 6.48 (superscript annotations as per Ballesteros-Weinstein nomenclature) suggest a distinct activation process. We characterized the interfaces of MRGPRX2 and the G i trimer, and mapped the residues associated with key single-nucleotide polymorphisms on both the ligand and G-protein interfaces of MRGPRX2. Collectively, our results provide a structural basis for the sensing of cationic allergens by MRGPRX2, potentially facilitating the rational design of therapies to prevent unwanted pseudoallergic reactions.
Organizational Affiliation:
Key Laboratory of Molecular Cardiovascular Science, Department of Physiology and Pathophysiology, School of Basic Medical Sciences, Peking University, Ministry of Education, Beijing, China.