Discovery of 8-(6-Methoxypyridin-3-yl)-1-(4-(piperazin-1-yl)-3-(trifluoromethyl)phenyl)-1,5-dihydro- 4H -[1,2,3]triazolo[4,5- c ]quinolin-4-one (CQ211) as a Highly Potent and Selective RIOK2 Inhibitor.
Ouyang, Y., Si, H., Zhu, C., Zhong, L., Ma, H., Li, Z., Xiong, H., Liu, T., Liu, Z., Zhang, Z., Zhang, Z.M., Cai, Q.(2022) J Med Chem 65: 7833-7842
- PubMed: 35584513
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00271
- Primary Citation of Related Structures:
7VBT - PubMed Abstract:
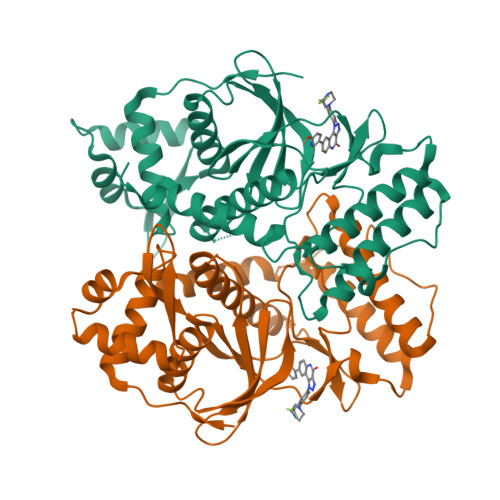
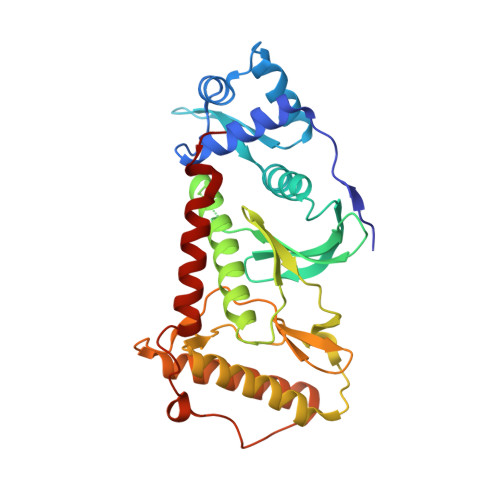
RIOK2 is an atypical kinase implicated in multiple human cancers. Although recent studies establish the role of RIOK2 in ribosome maturation and cell cycle progression, its biological functions remain poorly elucidated, hindering the potential to explore RIOK2 as a therapeutic target. Here, we report the discovery of CQ211 , the most potent and selective RIOK2 inhibitor reported so far. CQ211 displays a high binding affinity ( K d = 6.1 nM) and shows excellent selectivity to RIOK2 in both enzymatic and cellular studies. It also exhibits potent proliferation inhibition activity against multiple cancer cell lines and demonstrates promising in vivo efficacy in mouse xenograft models. The crystal structure of RIOK2- CQ211 sheds light on the molecular mechanism of inhibition and informs the subsequent optimization. The study provides a cell-active chemical probe for verifying RIOK2 functions, which may also serve as a leading molecule in the development of therapeutic RIOK2 inhibitors.
Organizational Affiliation:
College of Pharmacy, Jinan University, No. 601 Huangpu Avenue West, Guangzhou 510530, China.