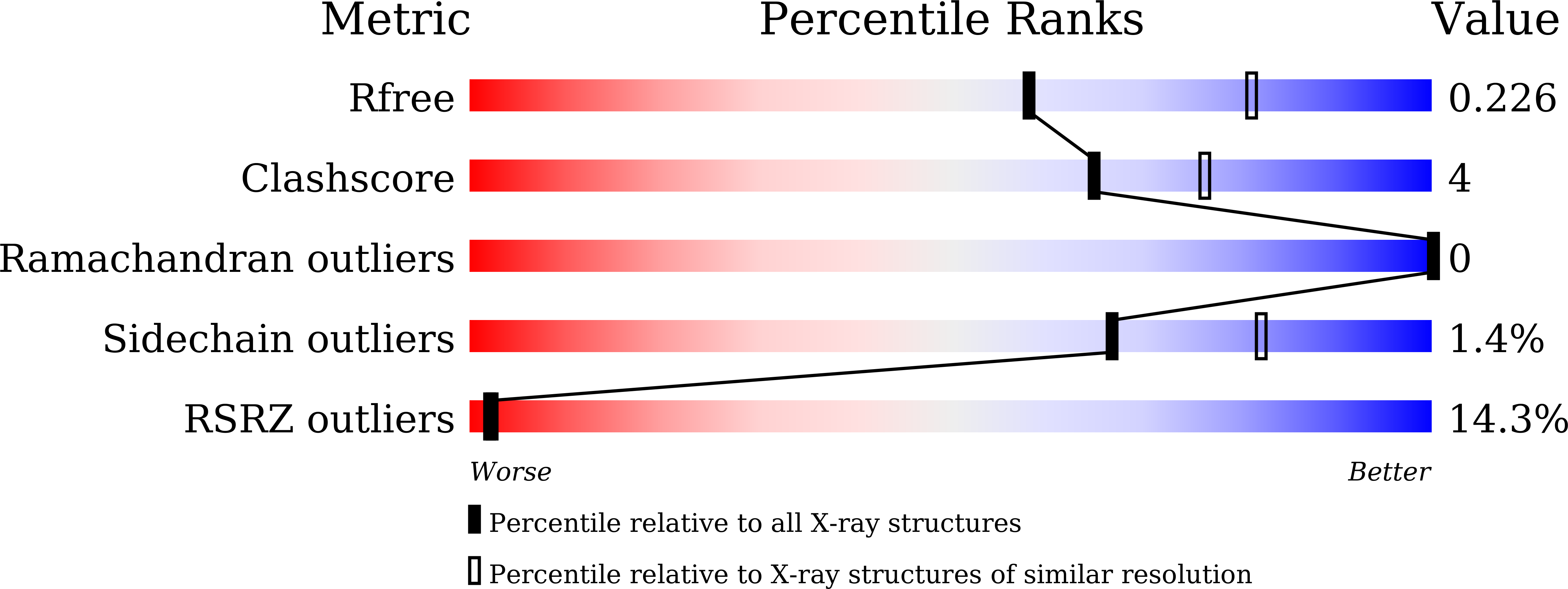
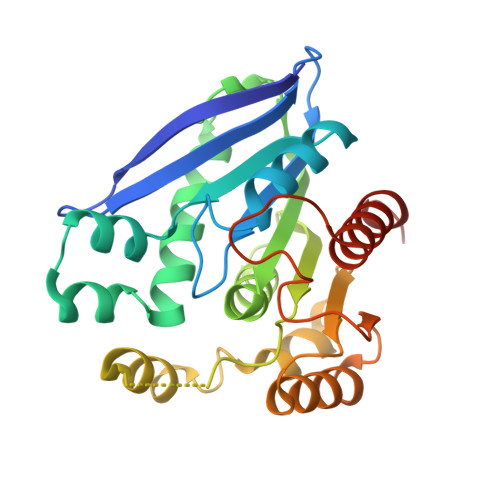
Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Son, J., Choi, W., Kim, H., Kim, M., Lee, J.H., Shin, S.C., Kim, H.W.(2023) IUCrJ 10: 220-232
- PubMed: 36862488
- DOI: https://doi.org/10.1107/S2052252523001562
- Primary Citation of Related Structures:
7V8U, 7V8V, 7V8W, 7V8X - PubMed Abstract:
PsEst3, a psychrophilic esterase obtained from Paenibacillus sp. R4, which was isolated from the permafrost of Alaska, exhibits relatively high activity at low temperatures. Here, crystal structures of PsEst3 complexed with various ligands were generated and studied at atomic resolution, and biochemical studies were performed to analyze the structure-function relationship of PsEst3. Certain unique characteristics of PsEst3 distinct from those of other classes of lipases/esterases were identified. Firstly, PsEst3 contains a conserved GHSRA/G pentapeptide sequence in the GxSxG motif around the nucleophilic serine. Additionally, it contains a conserved HGFR/K consensus sequence in the oxyanion hole, which is distinct from that in other lipase/esterase families, as well as a specific domain composition (for example a helix-turn-helix motif) and a degenerative lid domain that exposes the active site to the solvent. Secondly, the electrostatic potential of the active site in PsEst3 is positive, which may cause unintended binding of negatively charged chemicals in the active site. Thirdly, the last residue of the oxyanion hole-forming sequence, Arg44, separates the active site from the solvent by sealing the acyl-binding pocket, suggesting that PsEst3 is an enzyme that is customized to sense an unidentified substrate that is distinct from those of classical lipases/esterases. Collectively, this evidence strongly suggests that PsEst3 belongs to a distinct family of esterases.
Organizational Affiliation:
Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Republic of Korea.