Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Lim, X.X., Shu, B., Zhang, S., Tan, A.W.K., Ng, T.S., Lim, X.N., Chew, V.S., Shi, J., Screaton, G.R., Lok, S.M., Anand, G.S.(2021) Cell 184: 6067-6080.e13
- PubMed: 34852238
- DOI: https://doi.org/10.1016/j.cell.2021.11.009
- Primary Citation of Related Structures:
7V3F, 7V3G, 7V3H, 7V3I, 7V3J - PubMed Abstract:
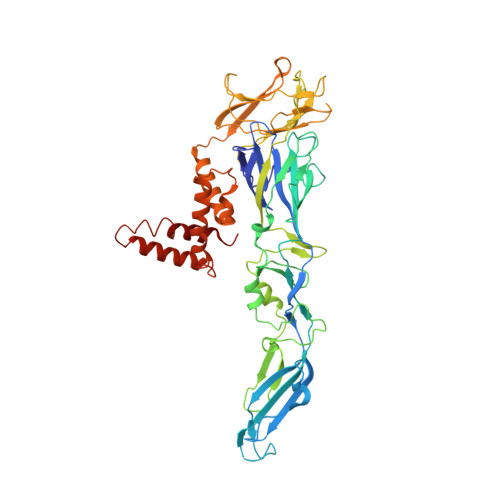
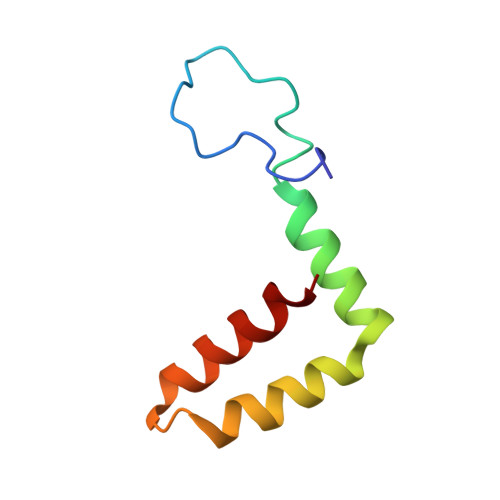
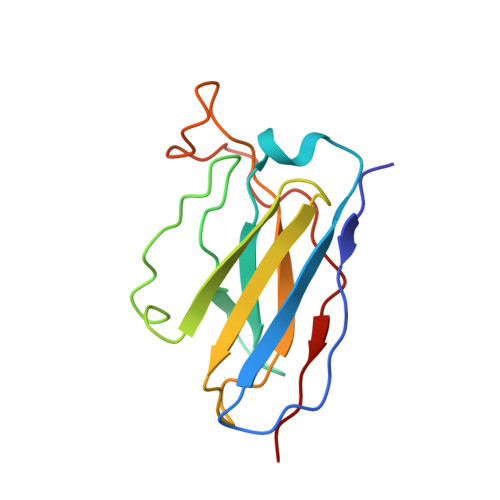
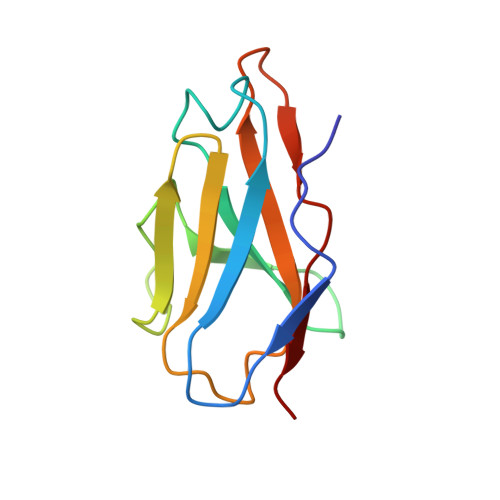
The human monoclonal antibody (HmAb) C10 potently cross-neutralizes Zika virus (ZIKV) and dengue virus. Analysis of antibody fragment (Fab) C10 interactions with ZIKV and dengue virus serotype 2 (DENV2) particles by cryoelectron microscopy (cryo-EM) and amide hydrogen/deuterium exchange mass spectrometry (HDXMS) shows that Fab C10 binding decreases overall ZIKV particle dynamics, whereas with DENV2, the same Fab causes increased dynamics. Testing of different Fab C10:DENV2 E protein molar ratios revealed that, at higher Fab ratios, especially at saturated concentrations, the Fab enhanced viral dynamics (detected by HDXMS), and observation under cryo-EM showed increased numbers of distorted particles. Our results suggest that Fab C10 stabilizes ZIKV but that with DENV2 particles, high Fab C10 occupancy promotes E protein dimer conformational changes leading to overall increased particle dynamics and distortion of the viral surface. This is the first instance of a broadly neutralizing antibody eliciting virus-specific increases in whole virus particle dynamics.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543, Singapore.