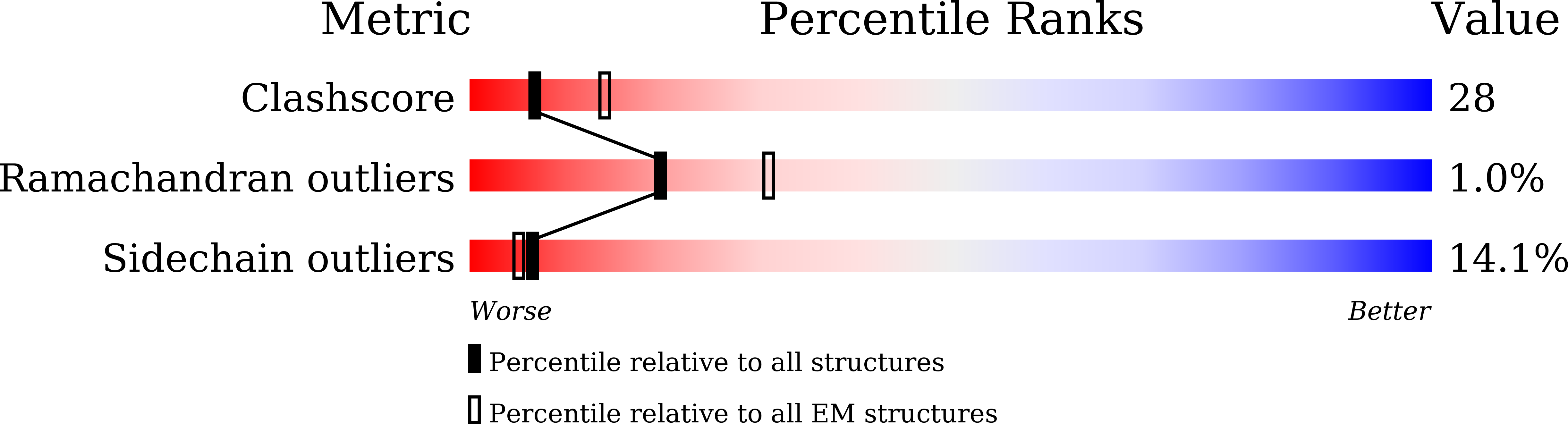TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Luo, J., Yang, Q., Zhang, X., Zhang, Y., Wan, L., Zhan, X., Zhou, Y., He, L., Li, D., Jin, D., Zhen, Y., Huang, J., Li, Y., Tao, L.(2022) Cell 185: 980-994.e15
- PubMed: 35303428
- DOI: https://doi.org/10.1016/j.cell.2022.02.010
- Primary Citation of Related Structures:
7V1N - PubMed Abstract:
The emergence of hypervirulent clade 2 Clostridioides difficile is associated with severe symptoms and accounts for >20% of global infections. TcdB is a dominant virulence factor of C. difficile, and clade 2 strains exclusively express two TcdB variants (TcdB2 and TcdB4) that use unknown receptors distinct from the classic TcdB. Here, we performed CRISPR/Cas9 screens for TcdB4 and identified tissue factor pathway inhibitor (TFPI) as its receptor. Using cryo-EM, we determined a complex structure of the full-length TcdB4 with TFPI, defining a common receptor-binding region for TcdB. Residue variations within this region divide major TcdB variants into 2 classes: one recognizes Frizzled (FZD), and the other recognizes TFPI. TFPI is highly expressed in the intestinal glands, and recombinant TFPI protects the colonic epithelium from TcdB2/4. These findings establish TFPI as a colonic crypt receptor for TcdB from clade 2 C. difficile and reveal new mechanisms for CDI pathogenesis.
Organizational Affiliation:
Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang 310024, China; Center for Infectious Disease Research, Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang 310024, China; Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang 310024, China.