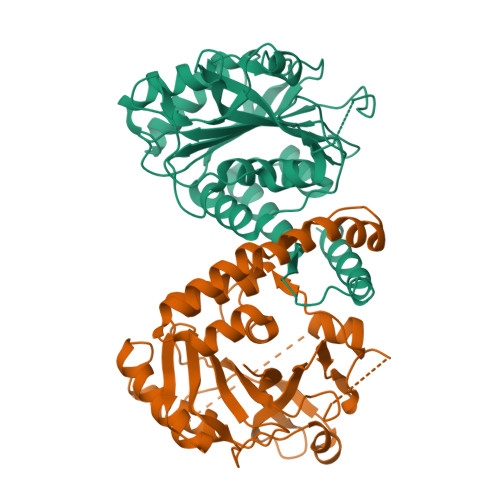
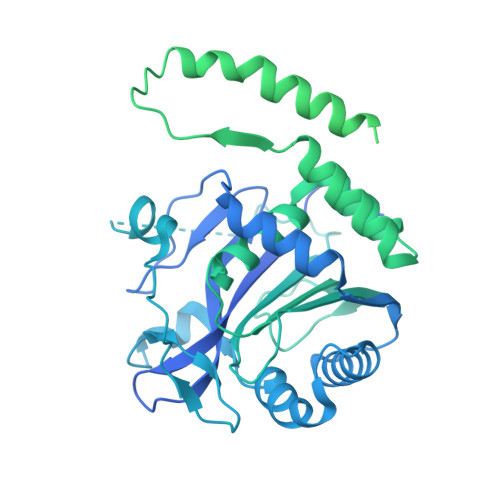
Architecture of the human erythrocyte ankyrin-1 complex.
Vallese, F., Kim, K., Yen, L.Y., Johnston, J.D., Noble, A.J., Cali, T., Clarke, O.B.(2022) Nat Struct Mol Biol 29: 706-718
- PubMed: 35835865
- DOI: https://doi.org/10.1038/s41594-022-00792-w
- Primary Citation of Related Structures:
7UZ3, 7UZE, 7UZQ, 7UZS, 7UZU, 7UZV, 7V07, 7V0K, 7V0M, 7V0Q, 7V0S, 7V0T, 7V0U, 7V0X, 7V0Y, 7V19, 8CRQ, 8CRR, 8CRT, 8CS9, 8CSL, 8CSV, 8CSW, 8CSX, 8CSY, 8CT2, 8CT3, 8CTE - PubMed Abstract:
The stability and shape of the erythrocyte membrane is provided by the ankyrin-1 complex, but how it tethers the spectrin-actin cytoskeleton to the lipid bilayer and the nature of its association with the band 3 anion exchanger and the Rhesus glycoproteins remains unknown. Here we present structures of ankyrin-1 complexes purified from human erythrocytes. We reveal the architecture of a core complex of ankyrin-1, the Rhesus proteins RhAG and RhCE, the band 3 anion exchanger, protein 4.2, glycophorin A and glycophorin B. The distinct T-shaped conformation of membrane-bound ankyrin-1 facilitates recognition of RhCE and, unexpectedly, the water channel aquaporin-1. Together, our results uncover the molecular details of ankyrin-1 association with the erythrocyte membrane, and illustrate the mechanism of ankyrin-mediated membrane protein clustering.
Organizational Affiliation:
Department of Anesthesiology, Columbia University Irving Medical Center, New York, NY, USA.