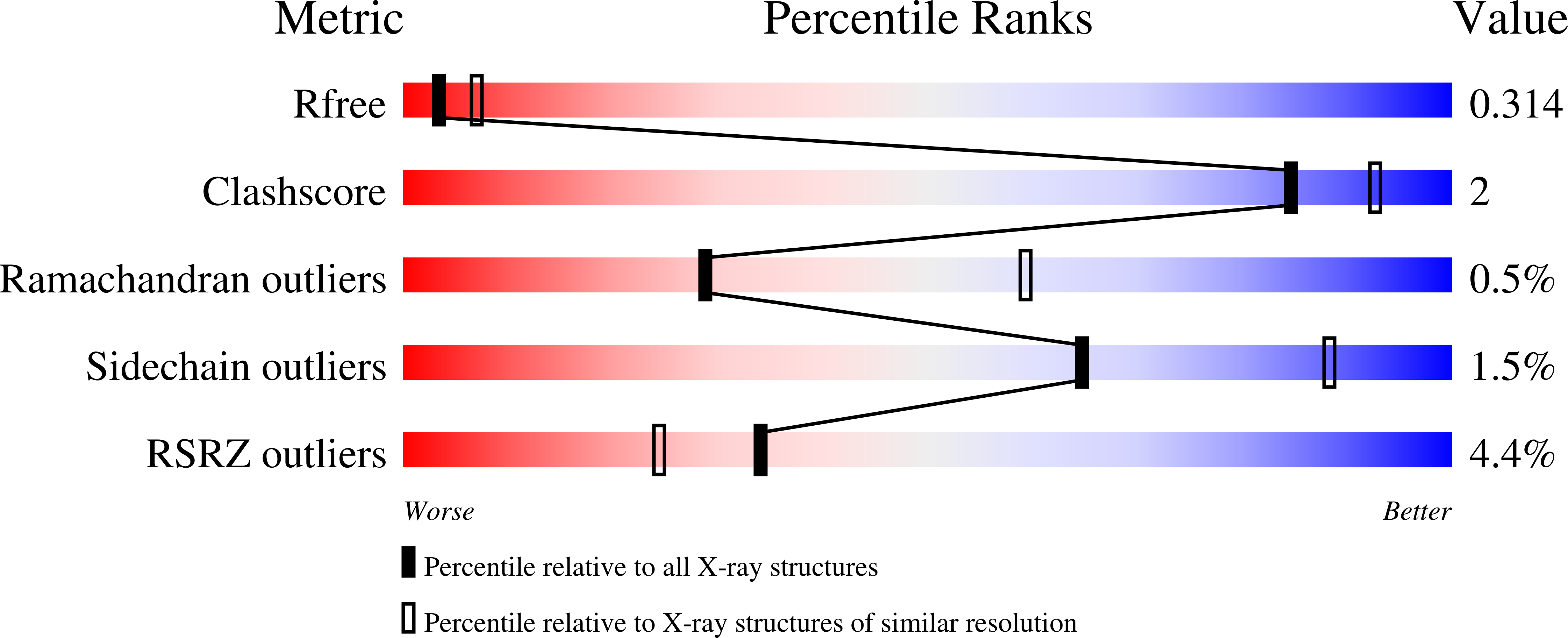
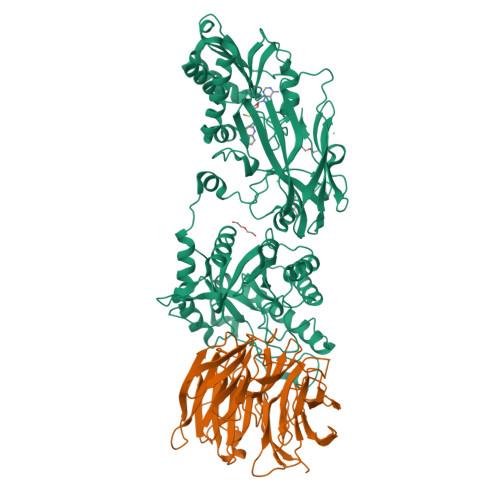
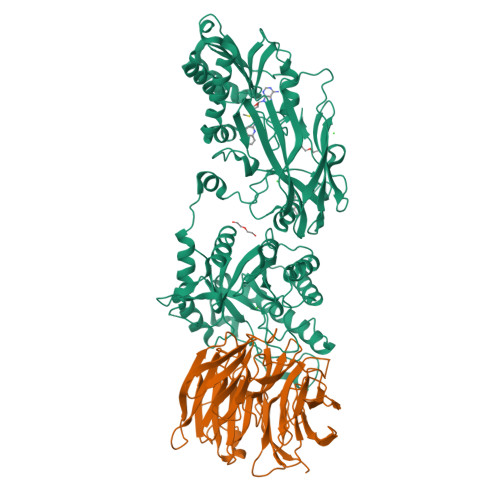
Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Smith, C.R., Kulyk, S., Ahmad, M.U.D., Arkhipova, V., Christensen, J.G., Gunn, R.J., Ivetac, A., Ketcham, J.M., Kuehler, J., Lawson, J.D., Thomas, N.C., Wang, X., Marx, M.A.(2022) RSC Med Chem 13: 1549-1564
- PubMed: 36545438
- DOI: https://doi.org/10.1039/d2md00163b
- Primary Citation of Related Structures:
7UY1, 7UYF, 7ZUP, 7ZUQ, 7ZUU, 7ZUY, 7ZV2, 7ZVL, 7ZVU, 8CSG, 8CTB - PubMed Abstract:
Here we describe the early stages of a fragment-based lead discovery (FBLD) project for a recently elucidated synthetic lethal target, the PRMT5/MTA complex, for the treatment of MTAP -deleted cancers. Starting with five fragment/PRMT5/MTA X-ray co-crystal structures, we employed a two-phase fragment elaboration process encompassing optimization of fragment hits and subsequent fragment growth to increase potency, assess synthetic tractability, and enable structure-based drug design. Two lead series were identified, one of which led to the discovery of the clinical candidate MRTX1719.
Organizational Affiliation:
Mirati Therapeutics San Diego California 92121 USA smithc@mirati.com kulyks@mirati.com.