RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Hao, Z., Gowder, M., Proshkin, S., Bharati, B.K., Epshtein, V., Svetlov, V., Shamovsky, I., Nudler, E.(2023) Cell 186: 2425
- PubMed: 37196657
- DOI: https://doi.org/10.1016/j.cell.2023.04.029
- Primary Citation of Related Structures:
7UWE, 7UWH - PubMed Abstract:
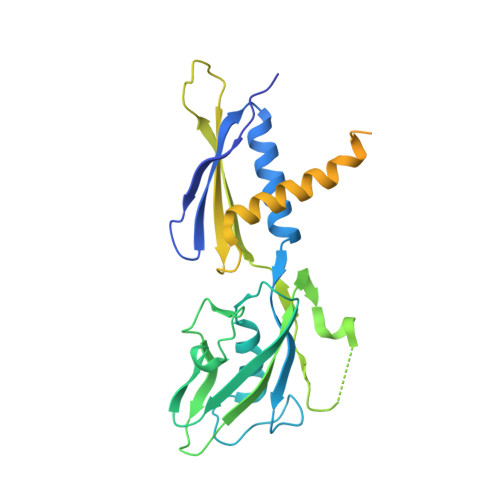
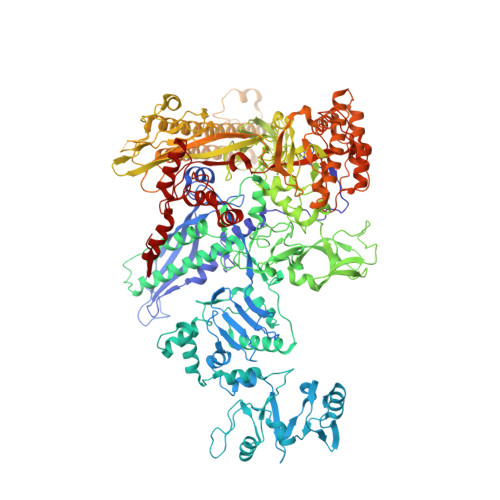
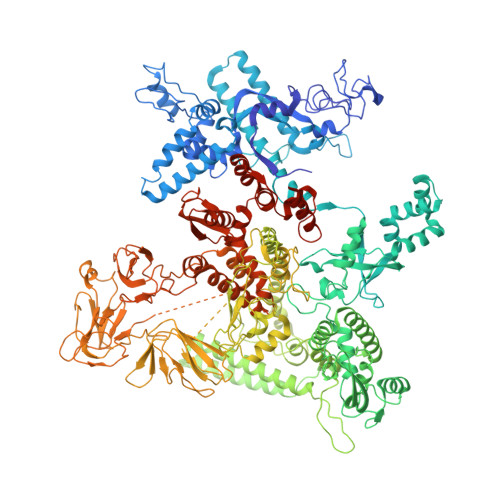
Ribonuclease HII (RNaseHII) is the principal enzyme that removes misincorporated ribonucleoside monophosphates (rNMPs) from genomic DNA. Here, we present structural, biochemical, and genetic evidence demonstrating that ribonucleotide excision repair (RER) is directly coupled to transcription. Affinity pull-downs and mass-spectrometry-assisted mapping of in cellulo inter-protein cross-linking reveal the majority of RNaseHII molecules interacting with RNA polymerase (RNAP) in E. coli. Cryoelectron microscopy structures of RNaseHII bound to RNAP during elongation, with and without the target rNMP substrate, show specific protein-protein interactions that define the transcription-coupled RER (TC-RER) complex in engaged and unengaged states. The weakening of RNAP-RNaseHII interactions compromises RER in vivo. The structure-functional data support a model where RNaseHII scans DNA in one dimension in search for rNMPs while "riding" the RNAP. We further demonstrate that TC-RER accounts for a significant fraction of repair events, thereby establishing RNAP as a surveillance "vehicle" for detecting the most frequently occurring replication errors.
Organizational Affiliation:
Department of Biochemistry and Molecular Pharmacology, New York University Grossman School of Medicine, New York, NY 10016, USA.