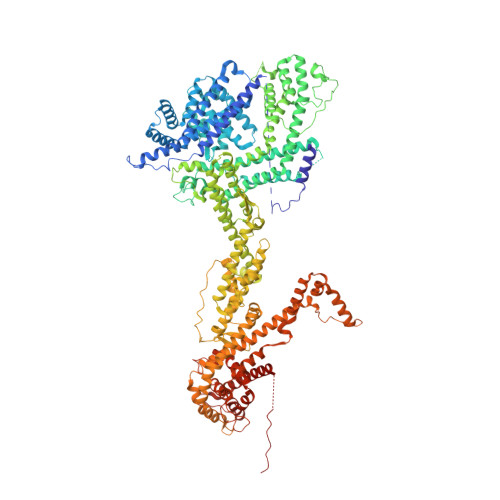
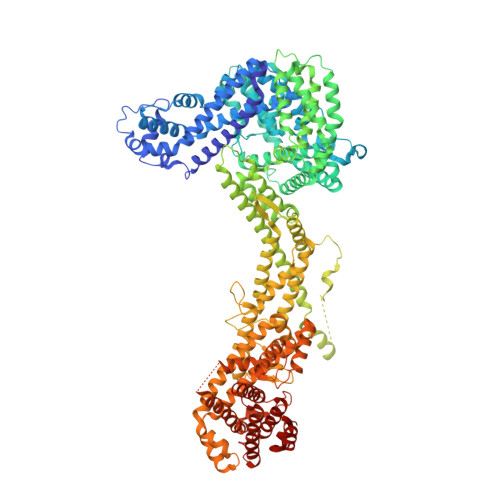
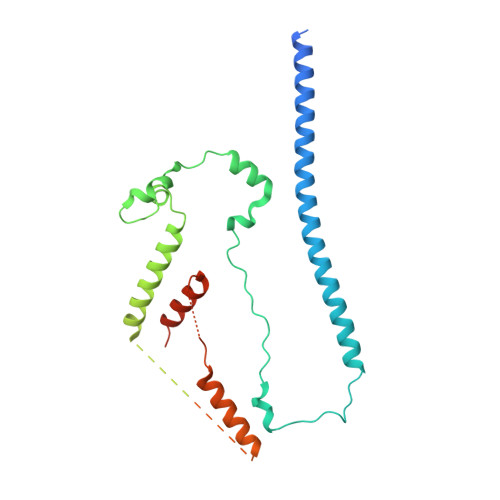
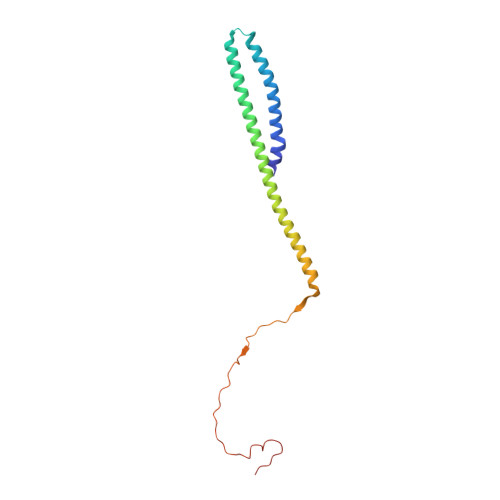
Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Ding, B., Yang, S., Schaks, M., Liu, Y., Brown, A.J., Rottner, K., Chowdhury, S., Chen, B.(2022) Nat Commun 13: 5444-5444
- PubMed: 36114192
- DOI: https://doi.org/10.1038/s41467-022-33174-3
- Primary Citation of Related Structures:
7USC, 7USD, 7USE - PubMed Abstract:
The Rho-family GTPase Rac1 activates the WAVE regulatory complex (WRC) to drive Arp2/3 complex-mediated actin polymerization in many essential processes. Rac1 binds to WRC at two distinct sites-the A and D sites. Precisely how Rac1 binds and how the binding triggers WRC activation remain unknown. Here we report WRC structures by itself, and when bound to single or double Rac1 molecules, at ~3 Å resolutions by cryogenic-electron microscopy. The structures reveal that Rac1 binds to the two sites by distinct mechanisms, and binding to the A site, but not the D site, drives WRC activation. Activation involves a series of unique conformational changes leading to the release of sequestered WCA (WH2-central-acidic) polypeptide, which stimulates the Arp2/3 complex to polymerize actin. Together with biochemical and cellular analyses, the structures provide a novel mechanistic understanding of how the Rac1-WRC-Arp2/3-actin signaling axis is regulated in diverse biological processes and diseases.
- Department of Biochemistry and Cell Biology, Stony Brook University, 100 Nicolls Road, Stony Brook, NY, 11794, USA.
Organizational Affiliation: