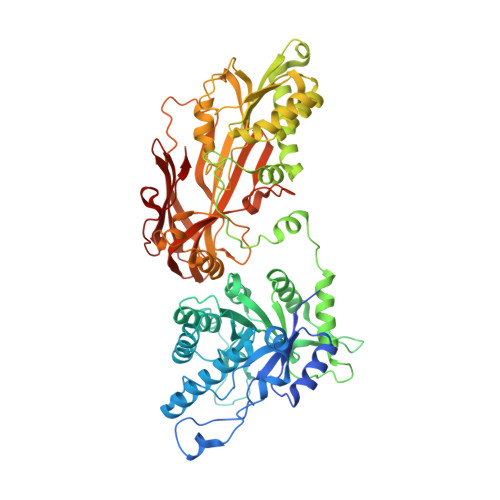
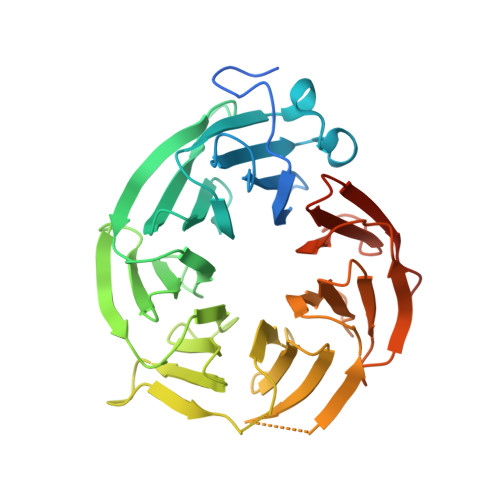
Design and evaluation of achiral, non-atropisomeric 4-(aminomethyl)phthalazin-1(2H)-one derivatives as novel PRMT5/MTA inhibitors.
Smith, C.R., Aranda, R., Christensen, J.G., Engstrom, L.D., Gunn, R.J., Ivetac, A., Ketcham, J.M., Kuehler, J., David Lawson, J., Marx, M.A., Olson, P., Thomas, N.C., Wang, X., Waters, L.M., Kulyk, S.(2022) Bioorg Med Chem 71: 116947-116947
- PubMed: 35926325
- DOI: https://doi.org/10.1016/j.bmc.2022.116947
- Primary Citation of Related Structures:
7UOH - PubMed Abstract:
MRTX1719 is an inhibitor of the PRMT5/MTA complex and recently entered clinical trials for the treatment of MTAP-deleted cancers. MRTX1719 is a class 3 atropisomeric compound that requires a chiral synthesis or a chiral separation step in its preparation. Here, we report the SAR and medicinal chemistry design strategy, supported by structural insights from X-ray crystallography, to discover a class 1 atropisomeric compound from the same series that does not require a chiral synthesis or a chiral separation step in its preparation.
Organizational Affiliation:
Mirati Therapeutics, San Diego, CA 92121, United States. Electronic address: smithc@mirati.com.