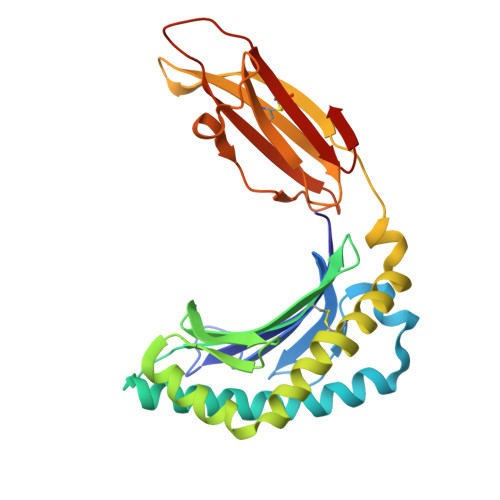
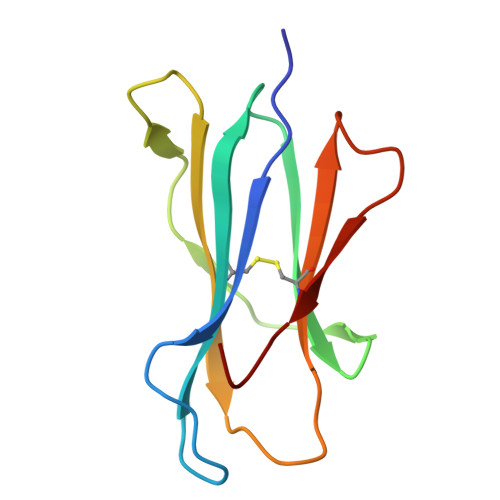
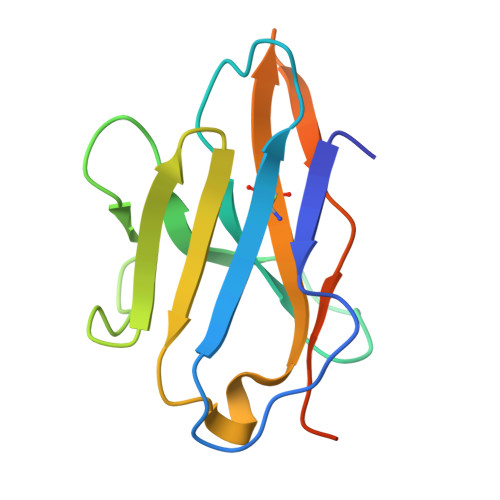
CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells.
Souter, M.N.T., Awad, W., Li, S., Pediongco, T.J., Meehan, B.S., Meehan, L.J., Tian, Z., Zhao, Z., Wang, H., Nelson, A., Le Nours, J., Khandokar, Y., Praveena, T., Wubben, J., Lin, J., Sullivan, L.C., Lovrecz, G.O., Mak, J.Y.W., Liu, L., Kostenko, L., Kedzierska, K., Corbett, A.J., Fairlie, D.P., Brooks, A.G., Gherardin, N.A., Uldrich, A.P., Chen, Z., Rossjohn, J., Godfrey, D.I., McCluskey, J., Pellicci, D.G., Eckle, S.B.G.(2022) J Exp Med 219
- PubMed: 36018322
- DOI: https://doi.org/10.1084/jem.20210828
- Primary Citation of Related Structures:
7UMG - PubMed Abstract:
Mucosal-associated invariant T (MAIT) cells detect microbial infection via recognition of riboflavin-based antigens presented by the major histocompatibility complex class I (MHC-I)-related protein 1 (MR1). Most MAIT cells in human peripheral blood express CD8αα or CD8αβ coreceptors, and the binding site for CD8 on MHC-I molecules is relatively conserved in MR1. Yet, there is no direct evidence of CD8 interacting with MR1 or the functional consequences thereof. Similarly, the role of CD8αα in lymphocyte function remains ill-defined. Here, using newly developed MR1 tetramers, mutated at the CD8 binding site, and by determining the crystal structure of MR1-CD8αα, we show that CD8 engaged MR1, analogous to how it engages MHC-I molecules. CD8αα and CD8αβ enhanced MR1 binding and cytokine production by MAIT cells. Moreover, the CD8-MR1 interaction was critical for the recognition of folate-derived antigens by other MR1-reactive T cells. Together, our findings suggest that both CD8αα and CD8αβ act as functional coreceptors for MAIT and other MR1-reactive T cells.
Organizational Affiliation:
Department of Microbiology and Immunology, The University of Melbourne at the Peter Doherty Institute for Infection and Immunity, Melbourne, Australia.