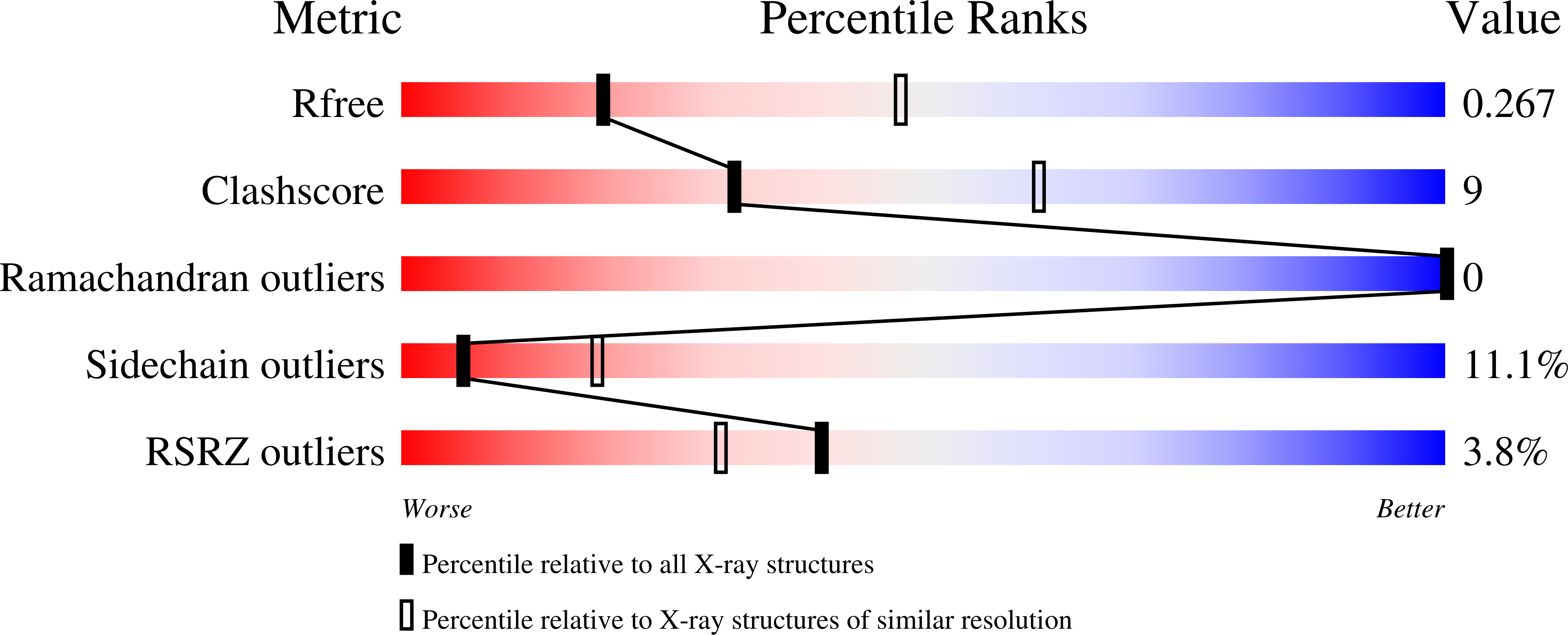
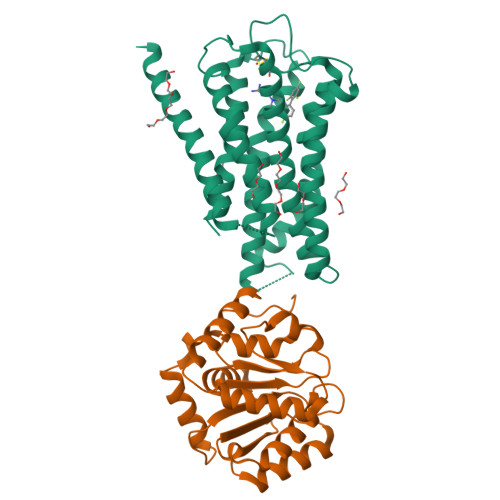
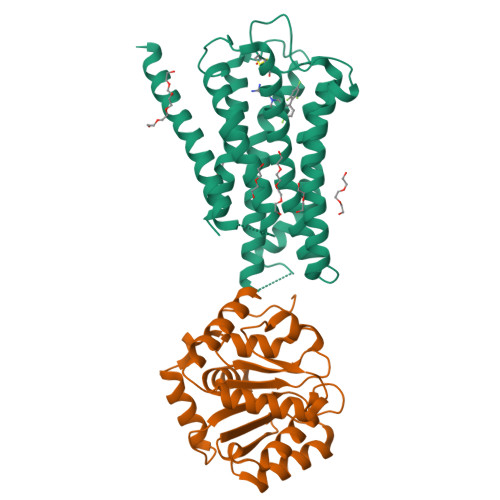
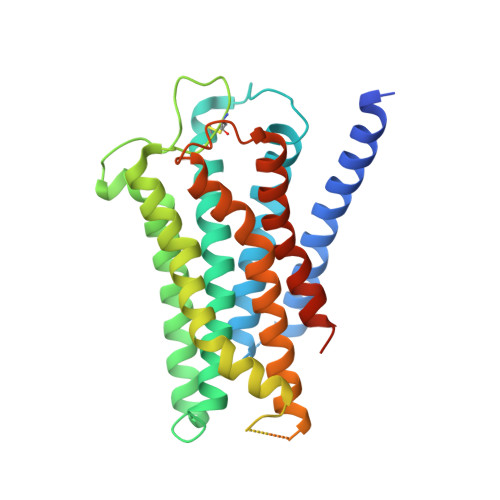
Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Zhang, S., Chen, H., Zhang, C., Yang, Y., Popov, P., Liu, J., Krumm, B.E., Cao, C., Kim, K., Xiong, Y., Katritch, V., Shoichet, B.K., Jin, J., Fay, J.F., Roth, B.L.(2022) Nat Struct Mol Biol 29: 677-687
- PubMed: 35835867
- DOI: https://doi.org/10.1038/s41594-022-00796-6
- Primary Citation of Related Structures:
7UM4, 7UM5, 7UM6, 7UM7 - PubMed Abstract:
Serotonin receptors are important targets for established therapeutics and drug development as they are expressed throughout the human body and play key roles in cell signaling. There are 12 serotonergic G protein-coupled receptor members encoded in the human genome, of which the 5-hydroxytryptamine (5-HT) 5A receptor (5-HT 5A R) is the least understood and lacks selective tool compounds. Here, we report four high-resolution (2.73-2.80 Å) structures of human 5-HT 5A Rs, including an inactive state structure bound to an antagonist AS2674723 by crystallization and active state structures bound to a partial agonist lisuride and two full agonists, 5-carboxamidotryptamine (5-CT) and methylergometrine, by cryo-EM. Leveraging the new structures, we developed a highly selective and potent antagonist for 5-HT 5A R. Collectively, these findings both enhance our understanding of this enigmatic receptor and provide a roadmap for structure-based drug discovery for 5-HT 5A R.
Organizational Affiliation:
Department of Pharmacology, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.