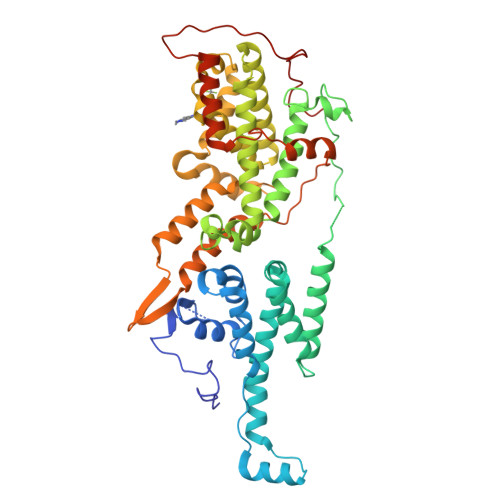
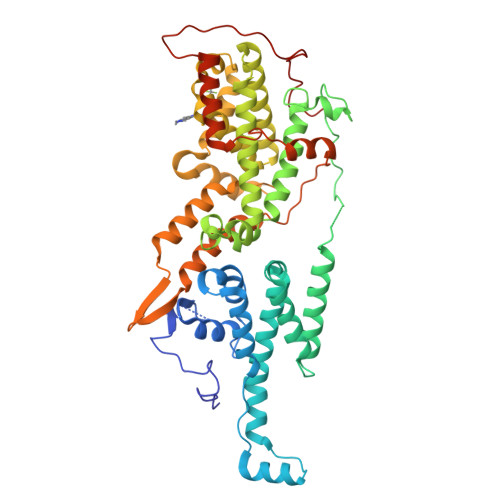
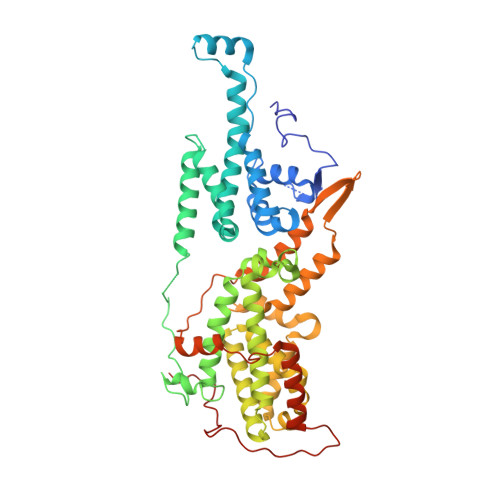
Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
Ketcham, J.M., Haling, J., Khare, S., Bowcut, V., Briere, D.M., Burns, A.C., Gunn, R.J., Ivetac, A., Kuehler, J., Kulyk, S., Laguer, J., Lawson, J.D., Moya, K., Nguyen, N., Rahbaek, L., Saechao, B., Smith, C.R., Sudhakar, N., Thomas, N.C., Vegar, L., Vanderpool, D., Wang, X., Yan, L., Olson, P., Christensen, J.G., Marx, M.A.(2022) J Med Chem 65: 9678-9690
- PubMed: 35833726
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00741
- Primary Citation of Related Structures:
7UKR, 7UKS - PubMed Abstract:
SOS1 is one of the major guanine nucleotide exchange factors that regulates the ability of KRAS to cycle through its "on" and "off" states. Disrupting the SOS1:KRAS G12C protein-protein interaction (PPI) can increase the proportion of GDP-loaded KRAS G12C , providing a strong mechanistic rationale for combining inhibitors of the SOS1:KRAS complex with inhibitors like MRTX849 that target GDP-loaded KRAS G12C . In this report, we detail the design and discovery of MRTX0902─a potent, selective, brain-penetrant, and orally bioavailable SOS1 binder that disrupts the SOS1:KRAS G12C PPI. Oral administration of MRTX0902 in combination with MRTX849 results in a significant increase in antitumor activity relative to that of either single agent, including tumor regressions in a subset of animals in the MIA PaCa-2 tumor mouse xenograft model.
Organizational Affiliation:
Mirati Therapeutics, 3545 Cray Court, San Diego, California 92121, United States.