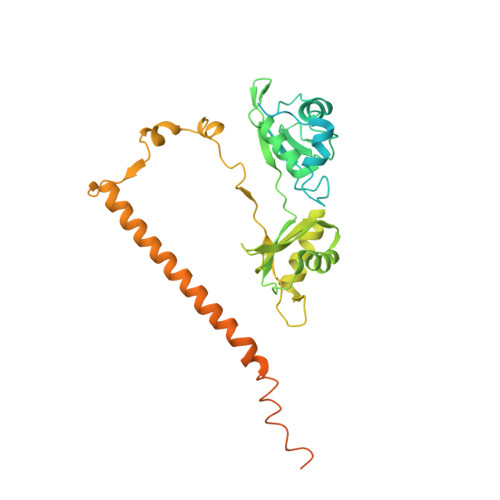
Insight into the Tumor Suppression Mechanism from the Structure of Human Polypyrimidine Splicing Factor (PSF/SFPQ) Complexed with a 30mer RNA from Murine Virus-like 30S Transcript-1.
Wang, J., Sachpatzidis, A., Christian, T.D., Lomakin, I.B., Garen, A., Konigsberg, W.H.(2022) Biochemistry 61: 1723-1734
- PubMed: 35998361
- DOI: https://doi.org/10.1021/acs.biochem.2c00192
- Primary Citation of Related Structures:
7UJ1, 7UK1 - PubMed Abstract:
Human polypyrimidine-binding splicing factor (PSF/SFPQ) is a tumor suppressor protein that regulates the gene expression of several proto-oncogenes and binds to the 5'-polyuridine negative-sense template (5'-PUN) of some RNA viruses. The activity of PSF is negatively regulated by long-noncoding RNAs, human metastasis associated in lung adenocarcinoma transcript-1 and murine virus-like 30S transcript-1 (VL30-1). PSF is a 707-amino acid protein that has a DNA-binding domain and two RNA recognition motifs (RRMs). Although the structure of the apo-truncated PSF is known, how PSF recognizes RNA remains elusive. Here, we report the 2.8 Å and 3.5 Å resolution crystal structures of a biologically active truncated construct of PSF (sPSF, consisting of residues 214-598) alone and in a complex with a 30mer fragment of VL30-1 RNA, respectively. The structure of the complex reveals how the 30mer RNA is recognized at two U-specific induced-fit binding pockets, located at the previously unrecognized domain-swapped, inter-subunit RRM1 (of the first subunit)-RRM2 (of the second subunit) interfaces that do not exist in the apo structure. Thus, the sPSF dimer appears to have two conformations in solution: one in a low-affinity state for RNA binding, as seen in the apo-structure, and the other in a high-affinity state for RNA binding, as seen in the sPSF-RNA complex. PSF undergoes an all or nothing transition between having two or no RNA-binding pockets. We predict that the RNA binds with a high degree of positive cooperativity. These structures provide an insight into a new regulatory mechanism that is likely involved in promoting malignancies and other human diseases.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, School of Medicine, Yale University, 333 Cedar Street, New Haven, Connecticut 06520-8114, USA.