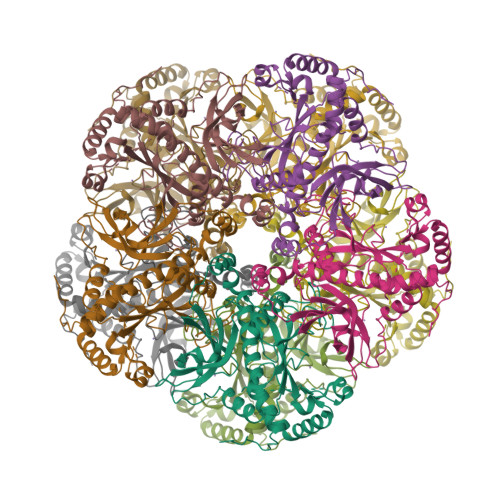
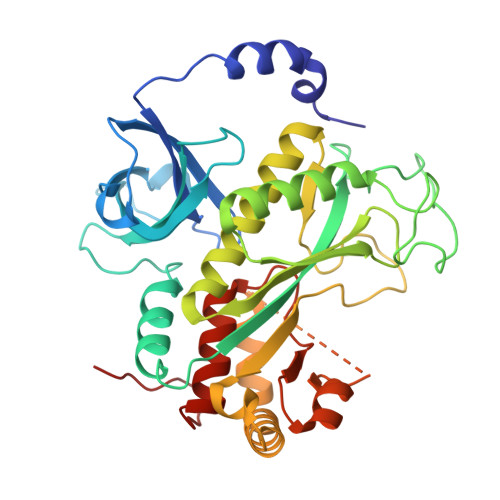
Toward structural-omics of the bovine retinal pigment epithelium.
Morgan, C.E., Zhang, Z., Miyagi, M., Golczak, M., Yu, E.W.(2022) Cell Rep 41: 111876-111876
- PubMed: 36577381
- DOI: https://doi.org/10.1016/j.celrep.2022.111876
- Primary Citation of Related Structures:
7U5H, 7U5I, 7U5J, 7U5K, 7U5L, 7U5M, 7U5N - PubMed Abstract:
The use of an integrated systems biology approach to investigate tissues and organs has been thought to be impracticable in the field of structural biology, where the techniques mainly focus on determining the structure of a particular biomacromolecule of interest. Here, we report the use of cryoelectron microscopy (cryo-EM) to define the composition of a raw bovine retinal pigment epithelium (RPE) lysate. From this sample, we simultaneously identify and solve cryo-EM structures of seven different RPE enzymes whose functions affect neurotransmitter recycling, iron metabolism, gluconeogenesis, glycolysis, axonal development, and energy homeostasis. Interestingly, dysfunction of these important proteins has been directly linked to several neurodegenerative disorders, including Huntington's disease, amyotrophic lateral sclerosis (ALS), Parkinson's disease, Alzheimer's disease, and schizophrenia. Our work underscores the importance of cryo-EM in facilitating tissue and organ proteomics at the atomic level.
Organizational Affiliation:
Department of Pharmacology, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA; Department of Chemistry, Thiel College, Greenville, PA 16125, USA.