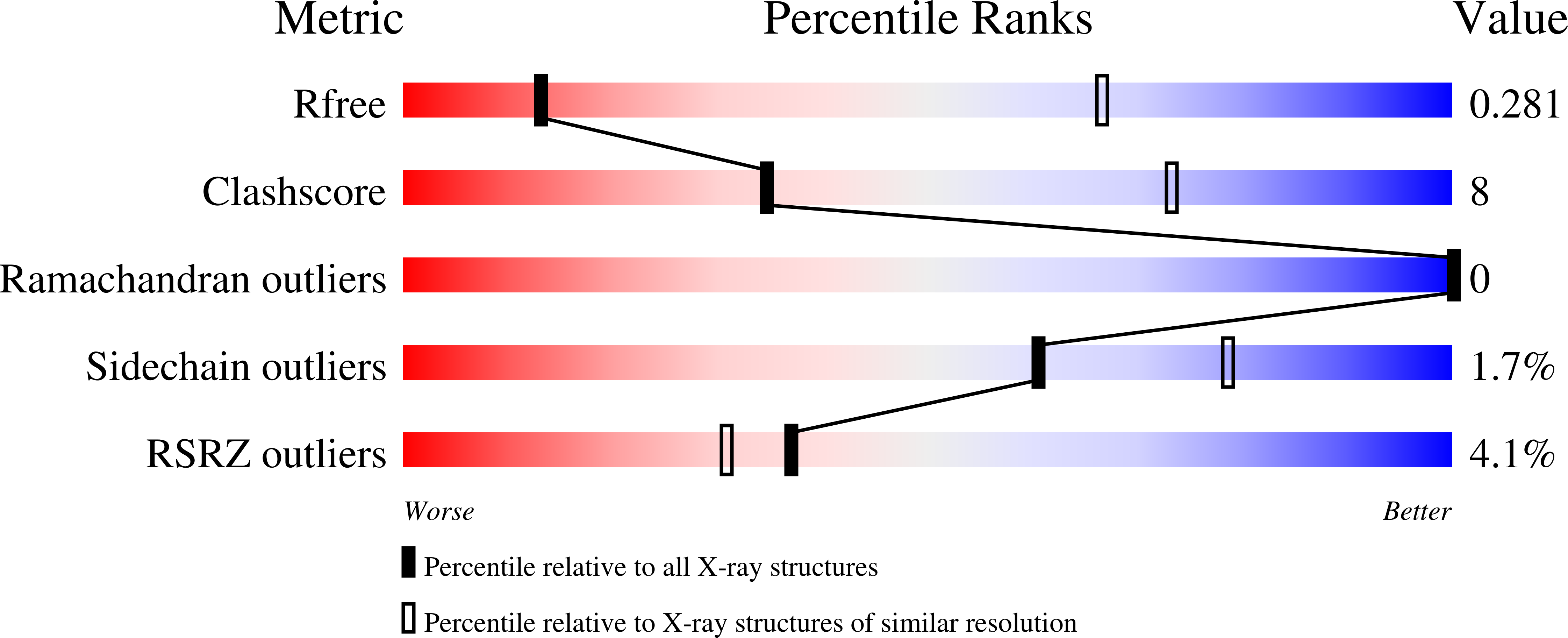
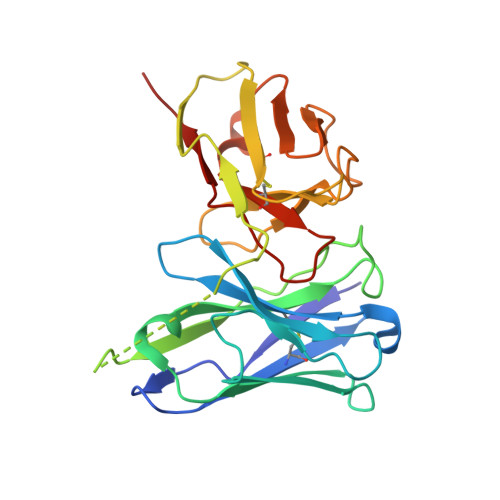
LAG3 ectodomain structure reveals functional interfaces for ligand and antibody recognition.
Ming, Q., Celias, D.P., Wu, C., Cole, A.R., Singh, S., Mason, C., Dong, S., Tran, T.H., Amarasinghe, G.K., Ruffell, B., Luca, V.C.(2022) Nat Immunol 23: 1031-1041
- PubMed: 35761082
- DOI: https://doi.org/10.1038/s41590-022-01238-7
- Primary Citation of Related Structures:
7TZ2, 7TZE, 7TZG, 7TZH - PubMed Abstract:
The immune checkpoint receptor lymphocyte activation gene 3 protein (LAG3) inhibits T cell function upon binding to major histocompatibility complex class II (MHC class II) or fibrinogen-like protein 1 (FGL1). Despite the emergence of LAG3 as a target for next-generation immunotherapies, we have little information describing the molecular structure of the LAG3 protein or how it engages cellular ligands. Here we determined the structures of human and murine LAG3 ectodomains, revealing a dimeric assembly mediated by Ig domain 2. Epitope mapping indicates that a potent LAG3 antagonist antibody blocks interactions with MHC class II and FGL1 by binding to a flexible 'loop 2' region in LAG3 domain 1. We also defined the LAG3-FGL1 interface by mapping mutations onto structures of LAG3 and FGL1 and established that FGL1 cross-linking induces the formation of higher-order LAG3 oligomers. These insights can guide LAG3-based drug development and implicate ligand-mediated LAG3 clustering as a mechanism for disrupting T cell activation.
Organizational Affiliation:
Department of Drug Discovery, Moffitt Cancer Center, Tampa, FL, USA.