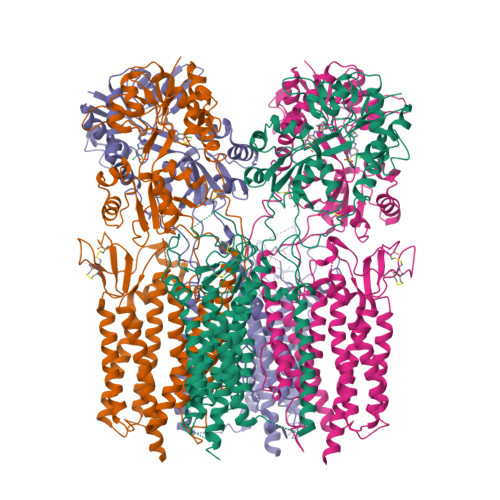
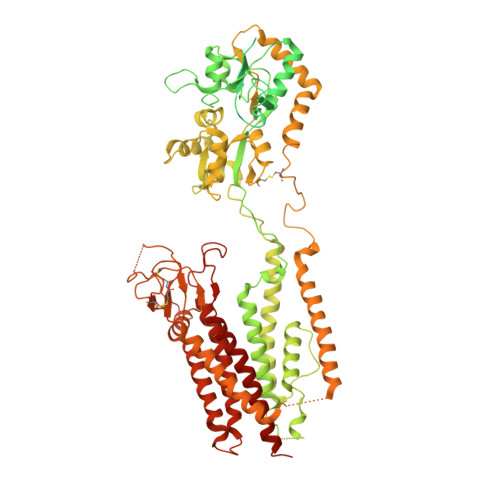
Opening of glutamate receptor channel to subconductance levels.
Yelshanskaya, M.V., Patel, D.S., Kottke, C.M., Kurnikova, M.G., Sobolevsky, A.I.(2022) Nature 605: 172-178
- PubMed: 35444281
- DOI: https://doi.org/10.1038/s41586-022-04637-w
- Primary Citation of Related Structures:
7TNJ, 7TNK, 7TNL, 7TNM, 7TNN, 7TNO, 7TNP - PubMed Abstract:
Ionotropic glutamate receptors (iGluRs) are tetrameric ligand-gated ion channels that open their pores in response to binding of the agonist glutamate 1-3 . An ionic current through a single iGluR channel shows up to four discrete conductance levels (O1-O4) 4-6 . Higher conductance levels have been associated with an increased number of agonist molecules bound to four individual ligand-binding domains (LBDs) 6-10 . Here we determine structures of a synaptic complex of AMPA-subtype iGluR and the auxiliary subunit γ2 in non-desensitizing conditions with various occupancy of the LBDs by glutamate. We show that glutamate binds to LBDs of subunits B and D only after it is already bound to at least the same number of LBDs that belong to subunits A and C. Our structures combined with single-channel recordings, molecular dynamics simulations and machine-learning analysis suggest that channel opening requires agonist binding to at least two LBDs. Conversely, agonist binding to all four LBDs does not guarantee maximal channel conductance and favours subconductance states O1 and O2, with O3 and O4 being rare and not captured structurally. The lack of subunit independence and low efficiency coupling of glutamate binding to channel opening underlie the gating of synaptic complexes to submaximal conductance levels, which provide a potential for upregulation of synaptic activity.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA.