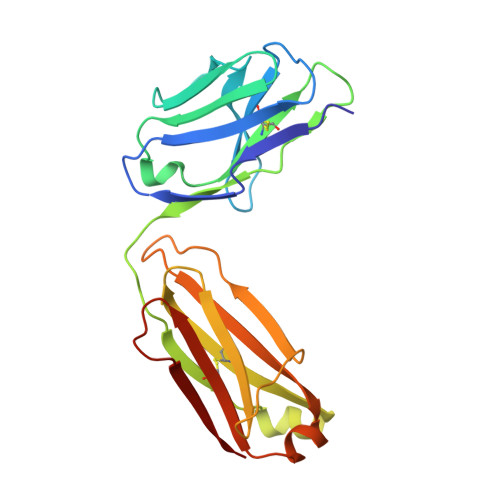
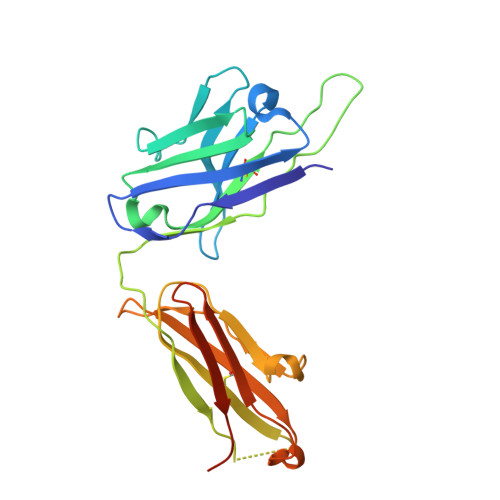
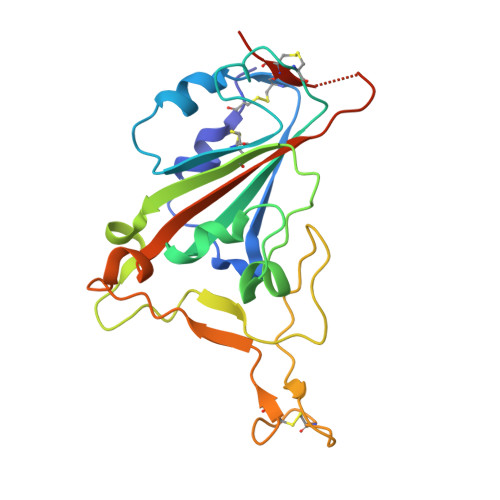
Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Rouet, R., Henry, J.Y., Johansen, M.D., Sobti, M., Balachandran, H., Langley, D.B., Walker, G.J., Lenthall, H., Jackson, J., Ubiparipovic, S., Mazigi, O., Schofield, P., Burnett, D.L., Brown, S.H.J., Martinello, M., Hudson, B., Gilroy, N., Post, J.J., Kelleher, A., Jack, H.M., Goodnow, C.C., Turville, S.G., Rawlinson, W.D., Bull, R.A., Stewart, A.G., Hansbro, P.M., Christ, D.(2023) Nat Commun 14: 687-687
- PubMed: 36755042
- DOI: https://doi.org/10.1038/s41467-023-36295-5
- Primary Citation of Related Structures:
7T5O, 7T72, 8DXT, 8DXU - PubMed Abstract:
Emerging variants of concern (VOCs) are threatening to limit the effectiveness of SARS-CoV-2 monoclonal antibodies and vaccines currently used in clinical practice; broadly neutralizing antibodies and strategies for their identification are therefore urgently required. Here we demonstrate that broadly neutralizing antibodies can be isolated from peripheral blood mononuclear cells of convalescent patients using SARS-CoV-2 receptor binding domains carrying epitope-specific mutations. This is exemplified by two human antibodies, GAR05, binding to epitope class 1, and GAR12, binding to a new epitope class 6 (located between class 3 and 5). Both antibodies broadly neutralize VOCs, exceeding the potency of the clinical monoclonal sotrovimab (S309) by orders of magnitude. They also provide prophylactic and therapeutic in vivo protection of female hACE2 mice against viral challenge. Our results indicate that exposure to SARS-CoV-2 induces antibodies that maintain broad neutralization against emerging VOCs using two unique strategies: either by targeting the divergent class 1 epitope in a manner resistant to VOCs (ACE2 mimicry, as illustrated by GAR05 and mAbs P2C-1F11/S2K14); or alternatively, by targeting rare and highly conserved epitopes, such as the new class 6 epitope identified here (as illustrated by GAR12). Our results provide guidance for next generation monoclonal antibody development and vaccine design.
Organizational Affiliation:
Garvan Institute of Medical Research, Sydney, NSW, Australia. r.rouet@garvan.org.au.