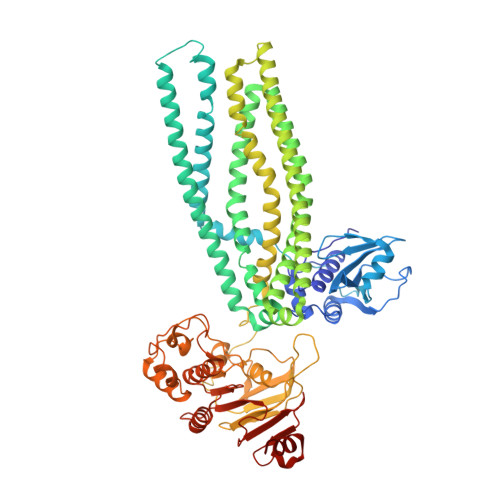
Structures of the peptidase-containing ABC transporter PCAT1 under equilibrium and nonequilibrium conditions.
Kieuvongngam, V., Chen, J.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35074919
- DOI: https://doi.org/10.1073/pnas.2120534119
- Primary Citation of Related Structures:
7T54, 7T55, 7T56, 7T57 - PubMed Abstract:
ATP-binding cassette (ABC) transporters are ubiquitous molecular pumps that transport a broad range of substrates across biological membranes. Although the structure and function of ABC transporters has been studied extensively, our understanding of their energetics and dynamics remains limited. Here, we present studies of the peptidase-containing ABC transporter 1 (PCAT1), a polypeptide processing and secretion ABC transporter that functions via the classic alternating access mechanism. PCAT1 is a homodimer containing two peptidase (PEP) domains, two transmembrane domains, and two nucleotide-binding domains (NBDs). Using cryo-electron microscopy, we analyzed the structures of wild-type PCAT1 under conditions that either prevent or permit ATP hydrolysis and observed two completely different conformational distributions. In the presence of ATP but absence of Mg 2+ , PCAT1 adopts an NBD-dimerized, outward-facing conformation. The two PEP domains are dissociated from the transporter core, preventing uncoupled substrate cleavage. The addition of Mg 2+ to promote ATP hydrolysis shifts the majority of the particles into NBD-separated, inward-facing conformations. Under this ATP turnover condition, only a small fraction of PCAT1 adopts the NBD-dimerized conformation. These data give rise to two mechanistic conclusions: 1) the ATP-bound, NBD-dimerized conformation is the lowest energy state, and 2) the rate-limiting step in the PCAT1 transport cycle is the formation of the NBD dimer. The thermodynamic conclusion is likely a general property shared by many ABC transporters. The kinetic bottleneck, however, varies from transporter to transporter.
Organizational Affiliation:
Laboratory of Membrane Biology and Biophysics, The Rockefeller University, New York, NY 10065.