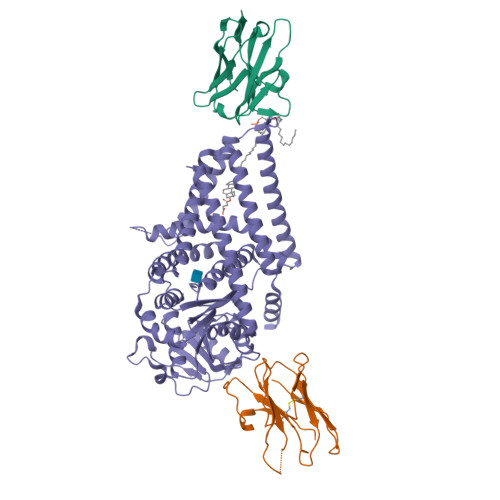
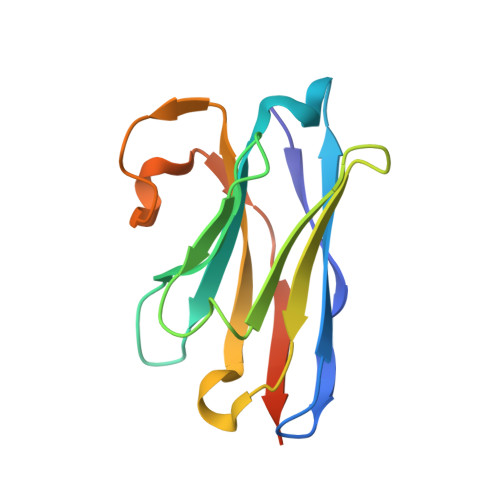
Structure, substrate recognition and initiation of hyaluronan synthase.
Maloney, F.P., Kuklewicz, J., Corey, R.A., Bi, Y., Ho, R., Mateusiak, L., Pardon, E., Steyaert, J., Stansfeld, P.J., Zimmer, J.(2022) Nature 604: 195-201
- PubMed: 35355017
- DOI: https://doi.org/10.1038/s41586-022-04534-2
- Primary Citation of Related Structures:
7SP6, 7SP7, 7SP8, 7SP9, 7SPA - PubMed Abstract:
Hyaluronan is an acidic heteropolysaccharide comprising alternating N-acetylglucosamine and glucuronic acid sugars that is ubiquitously expressed in the vertebrate extracellular matrix 1 . The high-molecular-mass polymer modulates essential physiological processes in health and disease, including cell differentiation, tissue homeostasis and angiogenesis 2 . Hyaluronan is synthesized by a membrane-embedded processive glycosyltransferase, hyaluronan synthase (HAS), which catalyses the synthesis and membrane translocation of hyaluronan from uridine diphosphate-activated precursors 3,4 . Here we describe five cryo-electron microscopy structures of a viral HAS homologue at different states during substrate binding and initiation of polymer synthesis. Combined with biochemical analyses and molecular dynamics simulations, our data reveal how HAS selects its substrates, hydrolyses the first substrate to prime the synthesis reaction, opens a hyaluronan-conducting transmembrane channel, ensures alternating substrate polymerization and coordinates hyaluronan inside its transmembrane pore. Our research suggests a detailed model for the formation of an acidic extracellular heteropolysaccharide and provides insights into the biosynthesis of one of the most abundant and essential glycosaminoglycans in the human body.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia School of Medicine, Charlottesville, VA, USA.