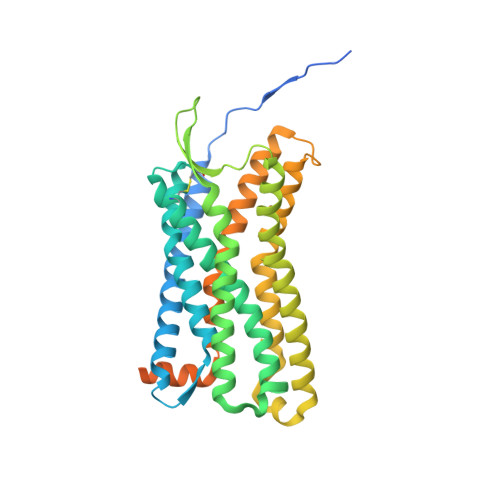
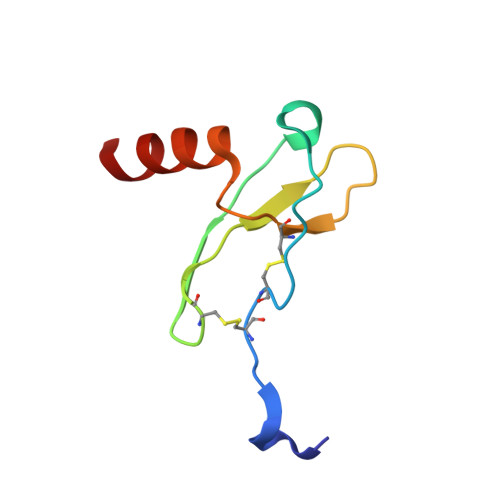
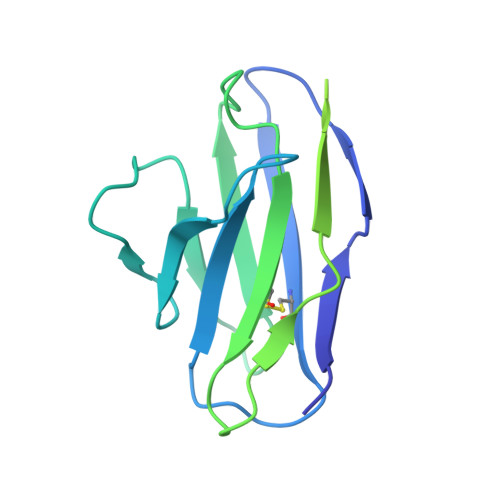
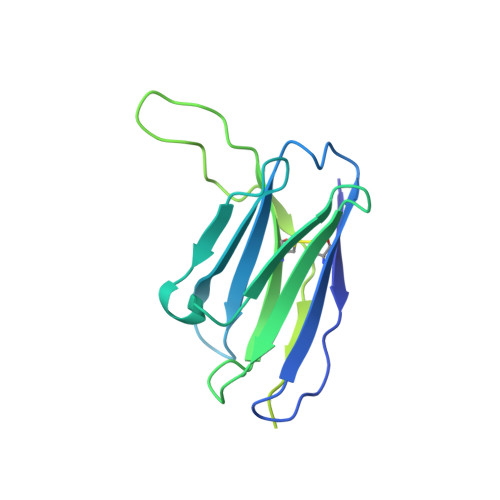
Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Yen, Y.C., Schafer, C.T., Gustavsson, M., Eberle, S.A., Dominik, P.K., Deneka, D., Zhang, P., Schall, T.J., Kossiakoff, A.A., Tesmer, J.J.G., Handel, T.M.(2022) Sci Adv 8: eabn8063-eabn8063
- PubMed: 35857509
- DOI: https://doi.org/10.1126/sciadv.abn8063
- Primary Citation of Related Structures:
7SK3, 7SK4, 7SK5, 7SK6, 7SK7, 7SK8, 7SK9 - PubMed Abstract:
Both CXC chemokine receptor 4 (CXCR4) and atypical chemokine receptor 3 (ACKR3) are activated by the chemokine CXCL12 yet evoke distinct cellular responses. CXCR4 is a canonical G protein-coupled receptor (GPCR), whereas ACKR3 is intrinsically biased for arrestin. The molecular basis for this difference is not understood. Here, we describe cryo-EM structures of ACKR3 in complex with CXCL12, a more potent CXCL12 variant, and a small-molecule agonist. The bound chemokines adopt an unexpected pose relative to those established for CXCR4 and observed in other receptor-chemokine complexes. Along with functional studies, these structures provide insight into the ligand-binding promiscuity of ACKR3, why it fails to couple to G proteins, and its bias toward β-arrestin. The results lay the groundwork for understanding the physiological interplay of ACKR3 with other GPCRs.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, IN, USA.