Structure of CDK4-Cyclin D3 bound to abemaciclib
Hilgers, M.T., Pelletier, L.A.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cyclin-dependent kinase 4 | 311 | Homo sapiens | Mutation(s): 1 Gene Names: CDK4 EC: 2.7.11.22 | 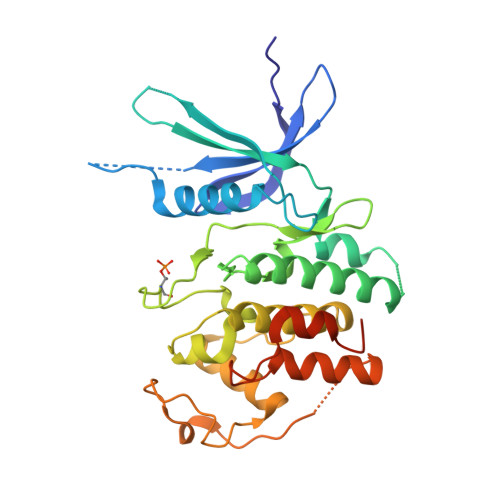 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P11802 (Homo sapiens) Explore P11802 Go to UniProtKB: P11802 | |||||
PHAROS: P11802 GTEx: ENSG00000135446 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P11802 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| G1/S-specific cyclin-D3 | 259 | Homo sapiens | Mutation(s): 0 Gene Names: CCND3 | 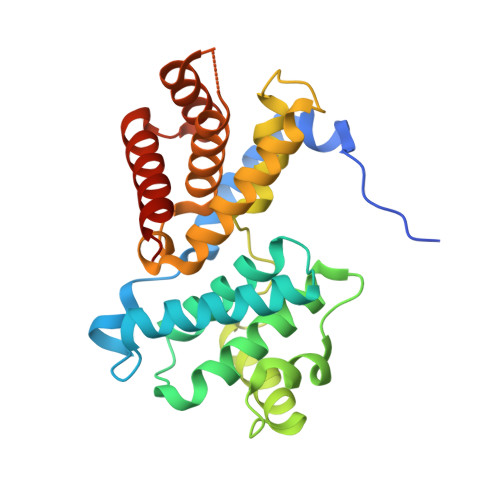 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P30281 (Homo sapiens) Explore P30281 Go to UniProtKB: P30281 | |||||
PHAROS: P30281 GTEx: ENSG00000112576 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30281 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 6ZV (Subject of Investigation/LOI) Query on 6ZV | C [auth A] | N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py
rimidin-2-amine C27 H32 F2 N8 UZWDCWONPYILKI-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| TPO Query on TPO | A | L-PEPTIDE LINKING | C4 H10 N O6 P |  | THR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 117.654 | α = 90 |
| b = 117.654 | β = 90 |
| c = 170.278 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| autoPROC | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | United States | NA |