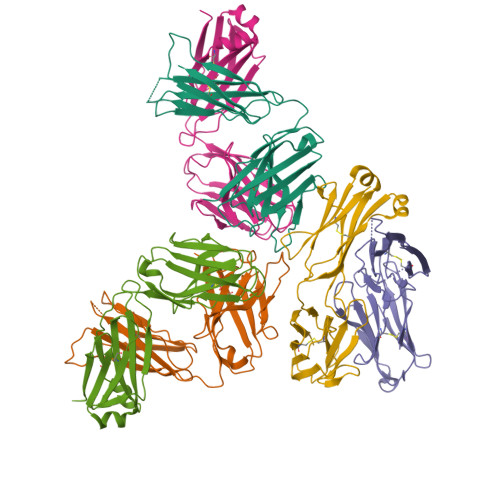
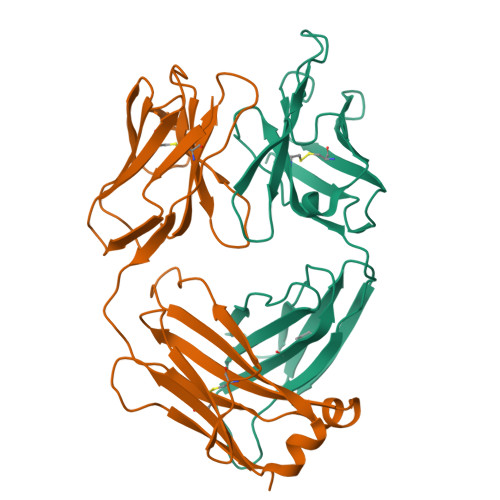
A native IgE in complex with profilin provides insights into allergen recognition and cross-reactivity.
Garcia-Ramirez, B., Mares-Mejia, I., Rodriguez-Hernandez, A., Cano-Sanchez, P., Torres-Larios, A., Ortega, E., Rodriguez-Romero, A.(2022) Commun Biol 5: 748-748
- PubMed: 35902770
- DOI: https://doi.org/10.1038/s42003-022-03718-w
- Primary Citation of Related Structures:
7SBD, 7SBG, 7SD2 - PubMed Abstract:
Allergies have become a rising health problem, where plentiful substances can trigger IgE-mediated allergies in humans. While profilins are considered minor allergens, these ubiquitous proteins are primary molecules involved in cross-reactivity and pollen-food allergy syndrome. Here we report the first crystal structures of murine Fab/IgE, with its chains naturally paired, in complex with the allergen profilin from Hevea brasiliensis (Hev b 8). The crystallographic models revealed that the IgE's six complementarity-determining regions (CDRs) interact with the allergen, comprising a rigid paratope-epitope surface of 926 Å 2 , which includes an extensive network of interactions. Interestingly, we also observed previously unreported flexibility at Fab/IgE's elbow angle, which did not influence the shape of the paratope. The Fab/IgE exhibits a high affinity for Hev b 8, even when using 1 M NaCl in BLI experiments. Finally, based on the encouraging cross-reactivity assays using two mutants of the maize profilin (Zea m 12), this antibody could be a promising tool in IgE engineering for diagnosis and research applications.
Organizational Affiliation:
Instituto de Química, Universidad Nacional Autónoma de México, Circuito Exterior, Cd. Universitaria, Coyoacán, Ciudad de México, 04510, Mexico.