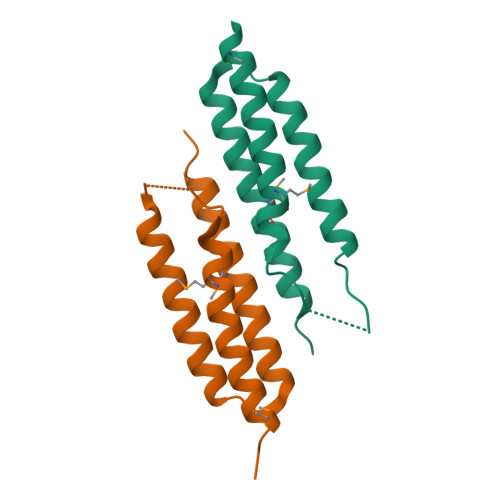
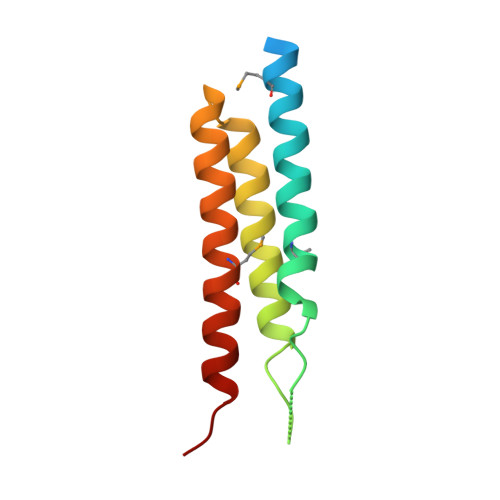
Group B Streptococcus Surface Protein beta : Structural Characterization of a Complement Factor H-Binding Motif and Its Contribution to Immune Evasion.
Xu, X., Lewis Marffy, A.L., Keightley, A., McCarthy, A.J., Geisbrecht, B.V.(2022) J Immunol 208: 1232-1247
- PubMed: 35110419
- DOI: https://doi.org/10.4049/jimmunol.2101078
- Primary Citation of Related Structures:
7S0R - PubMed Abstract:
The β protein from group B Streptococcus (GBS) is a ∼132-kDa, cell-surface exposed molecule that binds to multiple host-derived ligands, including complement factor H (FH). Many details regarding this interaction and its significance to immune evasion by GBS remain unclear. In this study, we identified a three-helix bundle domain within the C-terminal half of the B75KN region of β as the major FH-binding determinant and determined its crystal structure at 2.5 Å resolution. Analysis of this structure suggested a role in FH binding for a loop region connecting helices α1 and α2, which we confirmed by mutagenesis and direct binding studies. Using a combination of protein cross-linking and mass spectrometry, we observed that B75KN bound to complement control protein (CCP)3 and CCP4 domains of FH. Although this binding site lies within a complement regulatory region of FH, we determined that FH bound by β retained its decay acceleration and cofactor activities. Heterologous expression of β by Lactococcus lactis resulted in recruitment of FH to the bacterial surface and a significant reduction of C3b deposition following exposure to human serum. Surprisingly, we found that FH binding by β was not required for bacterial resistance to phagocytosis by neutrophils or killing of bacteria by whole human blood. However, loss of the B75KN region significantly diminished bacterial survival in both assays. Although our results show that FH recruited to the bacterial surface through a high-affinity interaction maintains key complement-regulatory functions, they raise questions about the importance of FH binding to immune evasion by GBS as a whole.
Organizational Affiliation:
Department of Biochemistry & Molecular Biophysics, Kansas State University, Manhattan, KS.