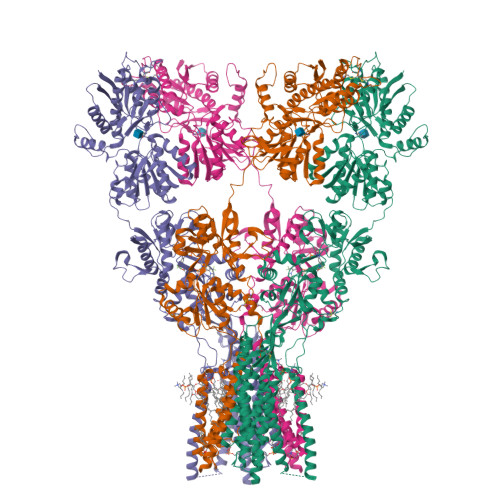
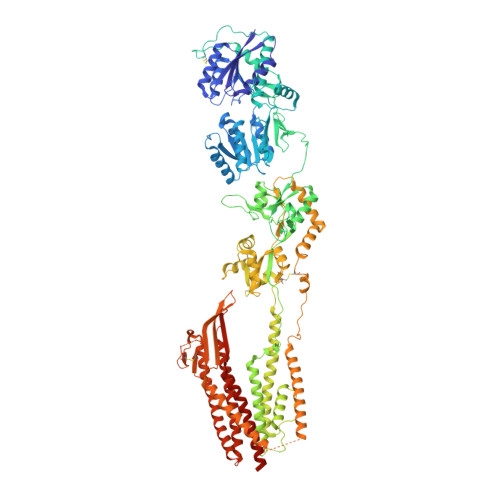
Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Klykov, O., Gangwar, S.P., Yelshanskaya, M.V., Yen, L., Sobolevsky, A.I.(2021) Mol Cell 81: 4771-4783.e7
- PubMed: 34678168
- DOI: https://doi.org/10.1016/j.molcel.2021.09.030
- Primary Citation of Related Structures:
7RYY, 7RYZ, 7RZ4, 7RZ5, 7RZ6, 7RZ7, 7RZ8, 7RZ9, 7RZA - PubMed Abstract:
AMPA receptors (AMPARs) mediate the majority of excitatory neurotransmission. Their surface expression, trafficking, gating, and pharmacology are regulated by auxiliary subunits. Of the two types of TARP auxiliary subunits, type I TARPs assume activating roles, while type II TARPs serve suppressive functions. We present cryo-EM structures of GluA2 AMPAR in complex with type II TARP γ5, which reduces steady-state currents, increases single-channel conductance, and slows recovery from desensitization. Regulation of AMPAR function depends on its ligand-binding domain (LBD) interaction with the γ5 head domain. GluA2-γ5 complex shows maximum stoichiometry of two TARPs per AMPAR tetramer, being different from type I TARPs but reminiscent of the auxiliary subunit GSG1L. Desensitization of both GluA2-GSG1L and GluA2-γ5 complexes is accompanied by rupture of LBD dimer interface, while GluA2-γ5 but not GluA2-GSG1L LBD dimers remain two-fold symmetric. Different structural architectures and desensitization mechanisms of complexes with auxiliary subunits endow AMPARs with broad functional capabilities.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, 650 West 168th Street, New York, NY 10032, USA.