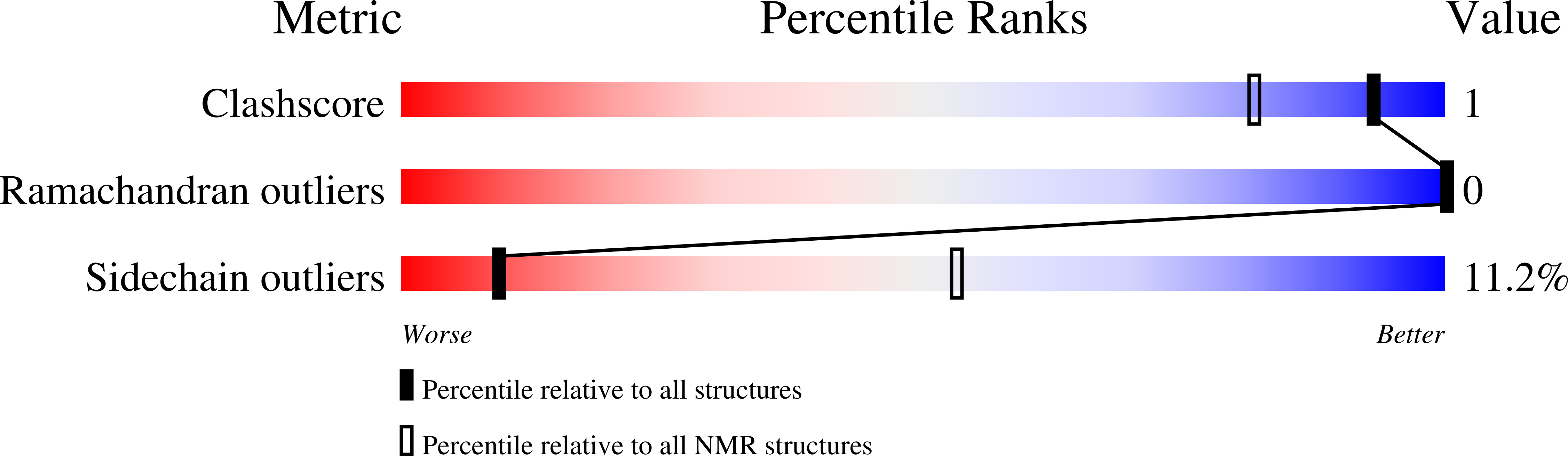A bivalent remipede toxin promotes calcium release via ryanodine receptor activation.
Maxwell, M.J., Thekkedam, C., Lamboley, C., Chin, Y.K., Crawford, T., Smith, J.J., Liu, J., Jia, X., Vetter, I., Laver, D.R., Launikonis, B.S., Dulhunty, A., Undheim, E.A.B., Mobli, M.(2023) Nat Commun 14: 1036-1036
- PubMed: 36823422
- DOI: https://doi.org/10.1038/s41467-023-36579-w
- Primary Citation of Related Structures:
7RZ3 - PubMed Abstract:
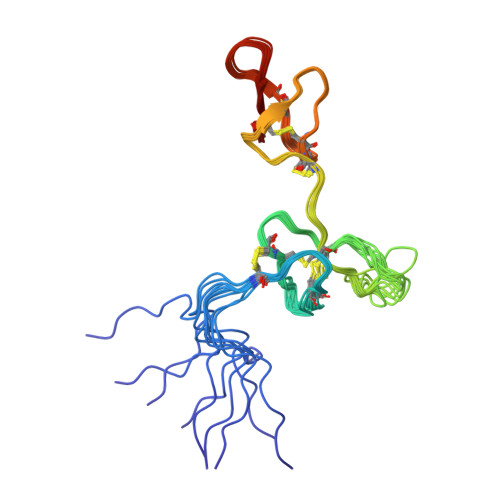
Multivalent ligands of ion channels have proven to be both very rare and highly valuable in yielding unique insights into channel structure and pharmacology. Here, we describe a bivalent peptide from the venom of Xibalbanus tulumensis, a troglobitic arthropod from the enigmatic class Remipedia, that causes persistent calcium release by activation of ion channels involved in muscle contraction. The high-resolution solution structure of φ-Xibalbin3-Xt3a reveals a tandem repeat arrangement of inhibitor-cysteine knot (ICK) domains previously only found in spider venoms. The individual repeats of Xt3a share sequence similarity with a family of scorpion toxins that target ryanodine receptors (RyR). Single-channel electrophysiology and quantification of released Ca 2+ stores within skinned muscle fibers confirm Xt3a as a bivalent RyR modulator. Our results reveal convergent evolution of RyR targeting toxins in remipede and scorpion venoms, while the tandem-ICK repeat architecture is an evolutionary innovation that is convergent with toxins from spider venoms.
Organizational Affiliation:
Centre for Advanced Imaging, Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, St. Lucia, QLD, 4072, Australia.