Structural insights into metazoan pretargeting GET complexes.
Keszei, A.F.A., Yip, M.C.J., Hsieh, T.C., Shao, S.(2021) Nat Struct Mol Biol 28: 1029-1037
- PubMed: 34887561
- DOI: https://doi.org/10.1038/s41594-021-00690-7
- Primary Citation of Related Structures:
7RU9, 7RUA, 7RUC - PubMed Abstract:
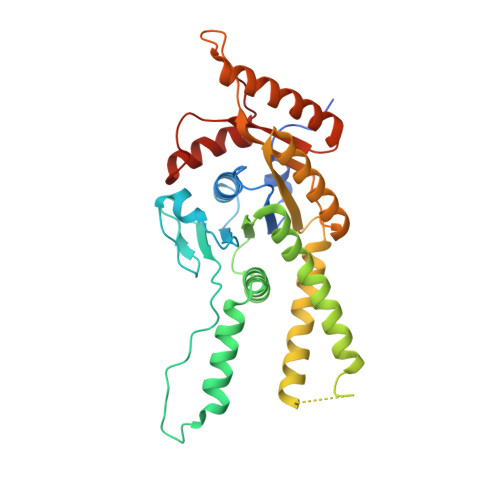
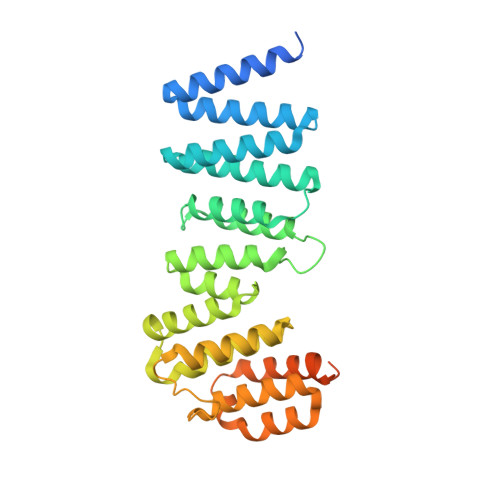
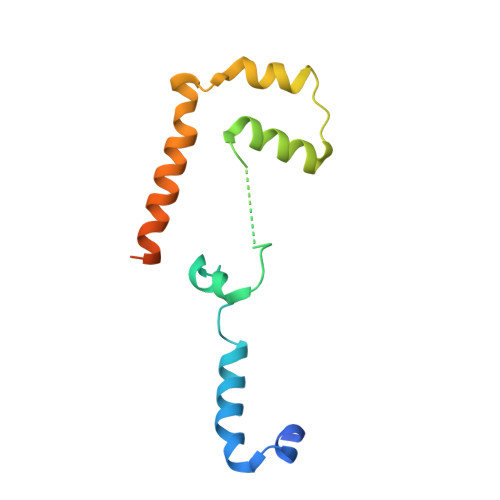
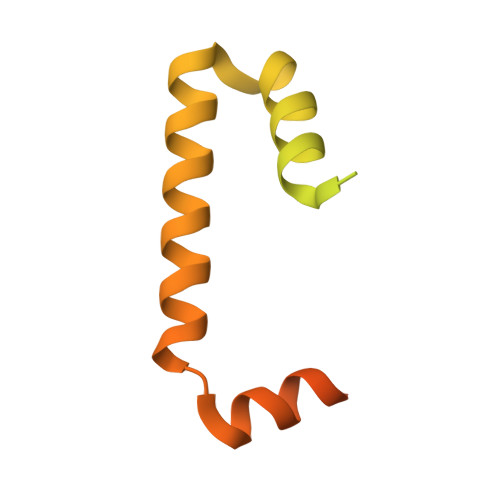
Close coordination between chaperones is essential for protein biosynthesis, including the delivery of tail-anchored (TA) proteins containing a single C-terminal transmembrane domain to the endoplasmic reticulum (ER) by the conserved GET pathway. For successful targeting, nascent TA proteins must be promptly chaperoned and loaded onto the cytosolic ATPase Get3 through a transfer reaction involving the chaperone SGTA and bridging factors Get4, Ubl4a and Bag6. Here, we report cryo-electron microscopy structures of metazoan pretargeting GET complexes at 3.3-3.6 Å. The structures reveal that Get3 helix 8 and the Get4 C terminus form a composite lid over the Get3 substrate-binding chamber that is opened by SGTA. Another interaction with Get4 prevents formation of Get3 helix 4, which links the substrate chamber and ATPase domain. Both interactions facilitate TA protein transfer from SGTA to Get3. Our findings show how the pretargeting complex primes Get3 for coordinated client loading and ER targeting.
Organizational Affiliation:
Department of Cell Biology, Harvard Medical School, Blavatnik Institute, Boston, MA, USA.