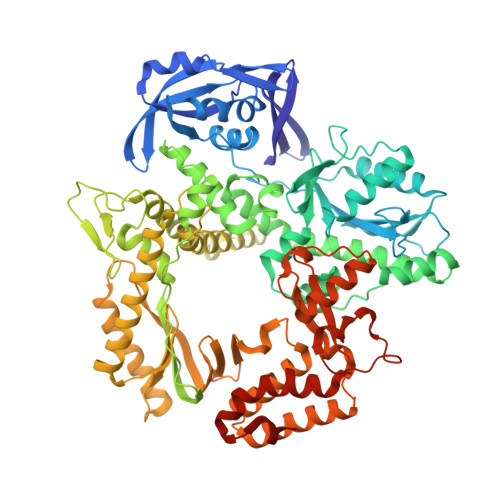
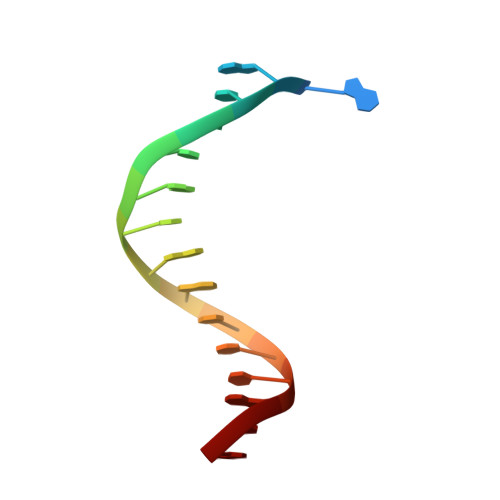
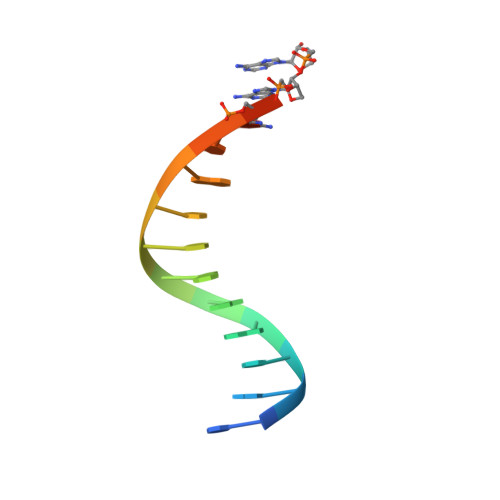
Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Hajjar, M., Chim, N., Liu, C., Herdewijn, P., Chaput, J.C.(2022) Nucleic Acids Res 50: 9663-9674
- PubMed: 36124684
- DOI: https://doi.org/10.1093/nar/gkac792
- Primary Citation of Related Structures:
7RSR, 7RSS, 7TQW - PubMed Abstract:
Xeno-nucleic acids (XNAs) are synthetic genetic polymers with backbone structures composed of non-ribose or non-deoxyribose sugars. Phosphonomethylthreosyl nucleic acid (pTNA), a type of XNA that does not base pair with DNA or RNA, has been suggested as a possible genetic material for storing synthetic biology information in cells. A critical step in this process is the synthesis of XNA episomes using laboratory-evolved polymerases to copy DNA information into XNA. Here, we investigate the polymerase recognition of pTNA nucleotides using X-ray crystallography to capture the post-catalytic complex of engineered polymerases following the sequential addition of two pTNA nucleotides onto the 3'-end of a DNA primer. High-resolution crystal structures reveal that the polymerase mediates Watson-Crick base pairing between the extended pTNA adducts and the DNA template. Comparative analysis studies demonstrate that the sugar conformation and backbone position of pTNA are structurally more similar to threose nucleic acid than DNA even though pTNA and DNA share the same six-atom backbone repeat length. Collectively, these findings provide new insight into the structural determinants that guide the enzymatic synthesis of an orthogonal genetic polymer, and may lead to the discovery of new variants that function with enhanced activity.
Organizational Affiliation:
Department of Pharmaceutical Sciences, University of California, Irvine, CA 92697-3958, USA.