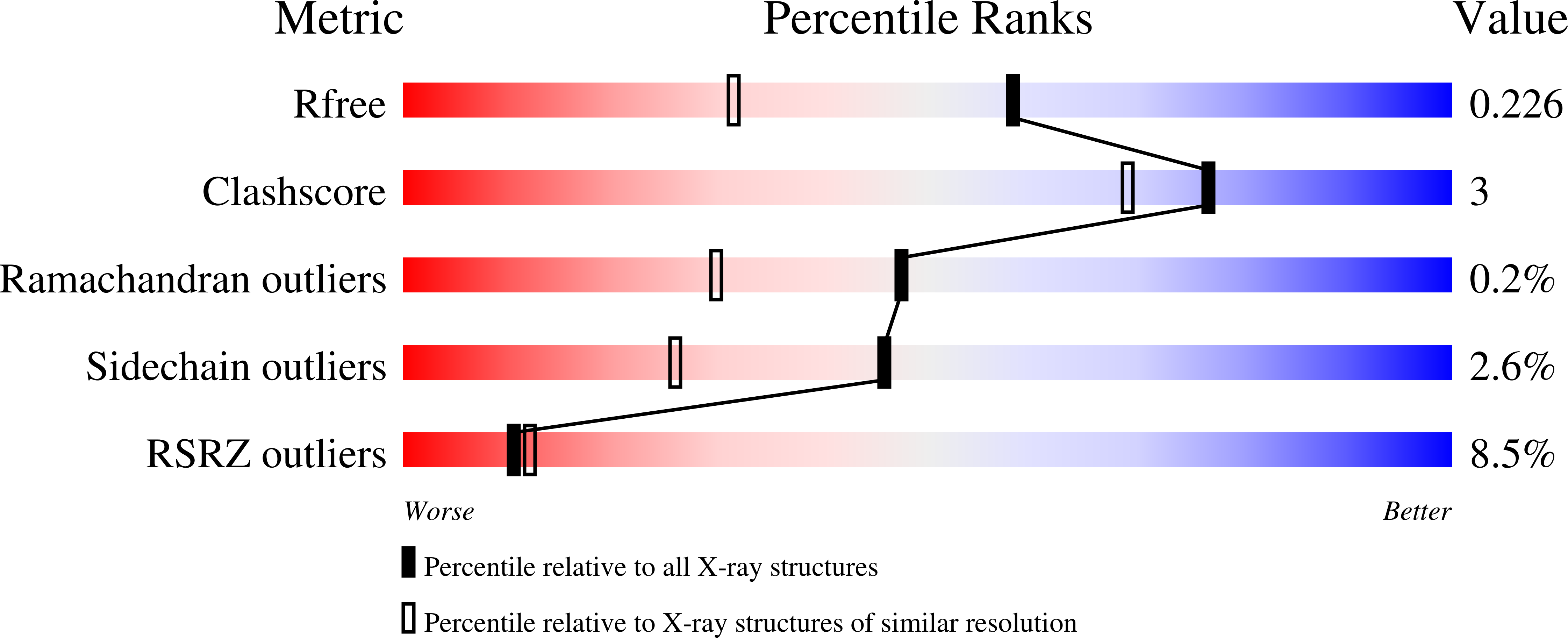
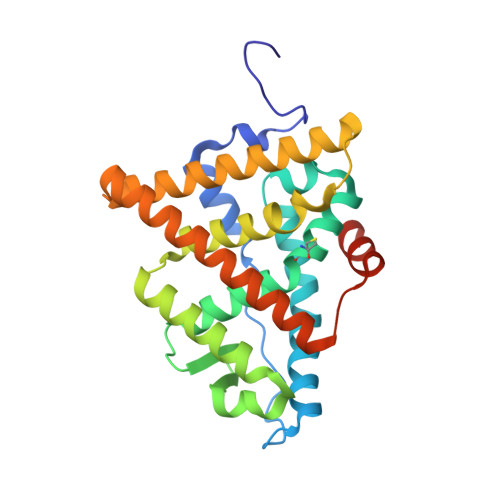
Dual-mechanism estrogen receptor inhibitors.
Min, J., Nwachukwu, J.C., Min, C.K., Njeri, J.W., Srinivasan, S., Rangarajan, E.S., Nettles, C.C., Sanabria Guillen, V., Ziegler, Y., Yan, S., Carlson, K.E., Hou, Y., Kim, S.H., Novick, S., Pascal, B.D., Houtman, R., Griffin, P.R., Izard, T., Katzenellenbogen, B.S., Katzenellenbogen, J.A., Nettles, K.W.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34452998
- DOI: https://doi.org/10.1073/pnas.2101657118
- Primary Citation of Related Structures:
7RRX, 7RRY, 7RRZ, 7RS0, 7RS1, 7RS2, 7RS3, 7RS4, 7RS7, 7RS8, 7RS9 - PubMed Abstract:
Efforts to improve estrogen receptor-α (ER)-targeted therapies in breast cancer have relied upon a single mechanism, with ligands having a single side chain on the ligand core that extends outward to determine antagonism of breast cancer growth. Here, we describe inhibitors with two ER-targeting moieties, one of which uses an alternate structural mechanism to generate full antagonism, freeing the side chain to independently determine other critical properties of the ligands. By combining two molecular targeting approaches into a single ER ligand, we have generated antiestrogens that function through new mechanisms and structural paradigms to achieve antagonism. These dual-mechanism ER inhibitors (DMERIs) cause alternate, noncanonical structural perturbations of the receptor ligand-binding domain (LBD) to antagonize proliferation in ER-positive breast cancer cells and in allele-specific resistance models. Our structural analyses with DMERIs highlight marked differences from current standard-of-care, single-mechanism antiestrogens. These findings uncover an enhanced flexibility of the ER LBD through which it can access nonconsensus conformational modes in response to DMERI binding, broadly and effectively suppressing ER activity.
Organizational Affiliation:
State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-Resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan 430062, China.