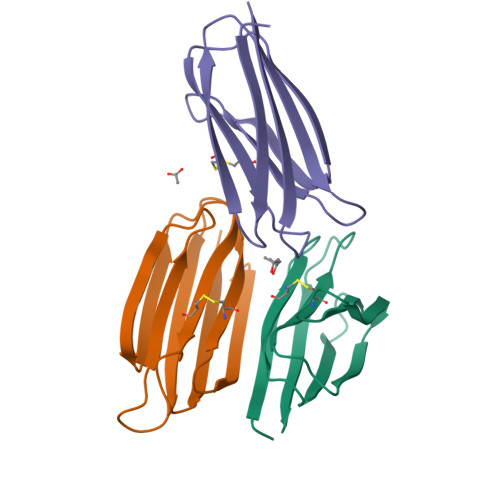Structural and thermodynamic analyses of human TMED1 (p24 gamma 1) Golgi dynamics.
Mota, D.C.A.M., Cardoso, I.A., Mori, R.M., Batista, M.R.B., Basso, L.G.M., Nonato, M.C., Costa-Filho, A.J., Mendes, L.F.S.(2022) Biochimie 192: 72-82
- PubMed: 34634369
- DOI: https://doi.org/10.1016/j.biochi.2021.10.002
- Primary Citation of Related Structures:
7RRM - PubMed Abstract:
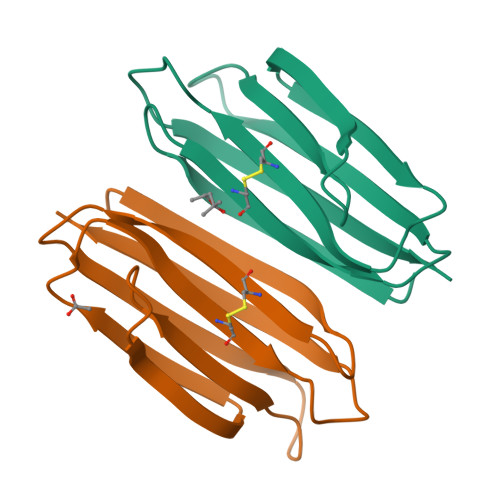
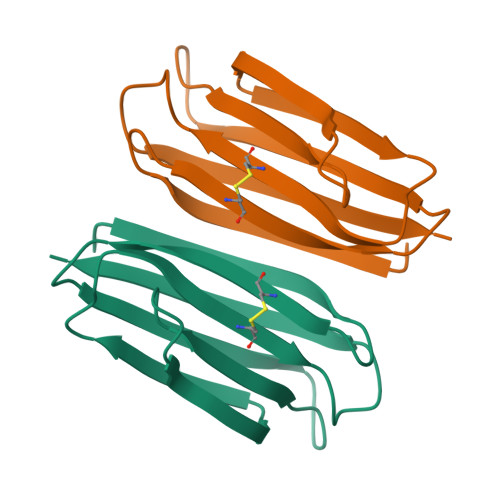
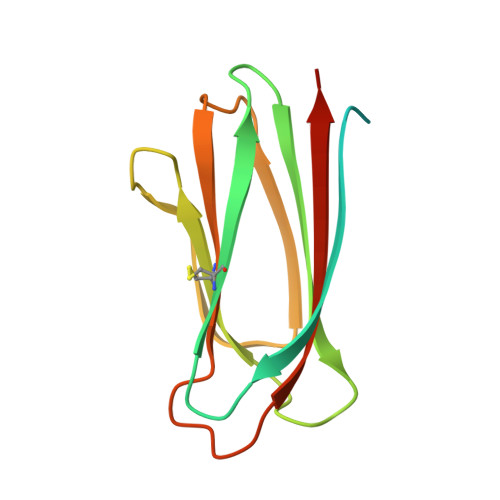
The transmembrane emp24 domain-containing (TMED) proteins, also called p24 proteins, are members of a family of sorting receptors present in all representatives of the Eukarya and abundantly present in all subcompartments of the early secretory pathway, namely the endoplasmic reticulum (ER), the Golgi, and the intermediate compartment. Although essential during the bidirectional transport between the ER and the Golgi, there is still a lack of information regarding the TMED's structure across different subfamilies. Besides, although the presence of a TMED homo-oligomerization was suggested previously based on crystallographic contacts observed for the isolated Golgi Dynamics (GOLD) domain, no further analyses of its presence in solution were done. Here, we describe the first high-resolution structure of a TMED1 GOLD representative and its biophysical characterization in solution. The crystal structure showed a dimer formation that is also present in solution in a salt-dependent manner, suggesting that the GOLD domain can form homodimers in solution even in the absence of the TMED1 coiled-coil region. A molecular dynamics description of the dimer stabilization, with a phylogenetic analysis of the residues important for the oligomerization and a model for the orientation towards the lipid membrane, are also presented.
Organizational Affiliation:
Laboratório de Biofísica Molecular, Departamento de Física, Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brazil.