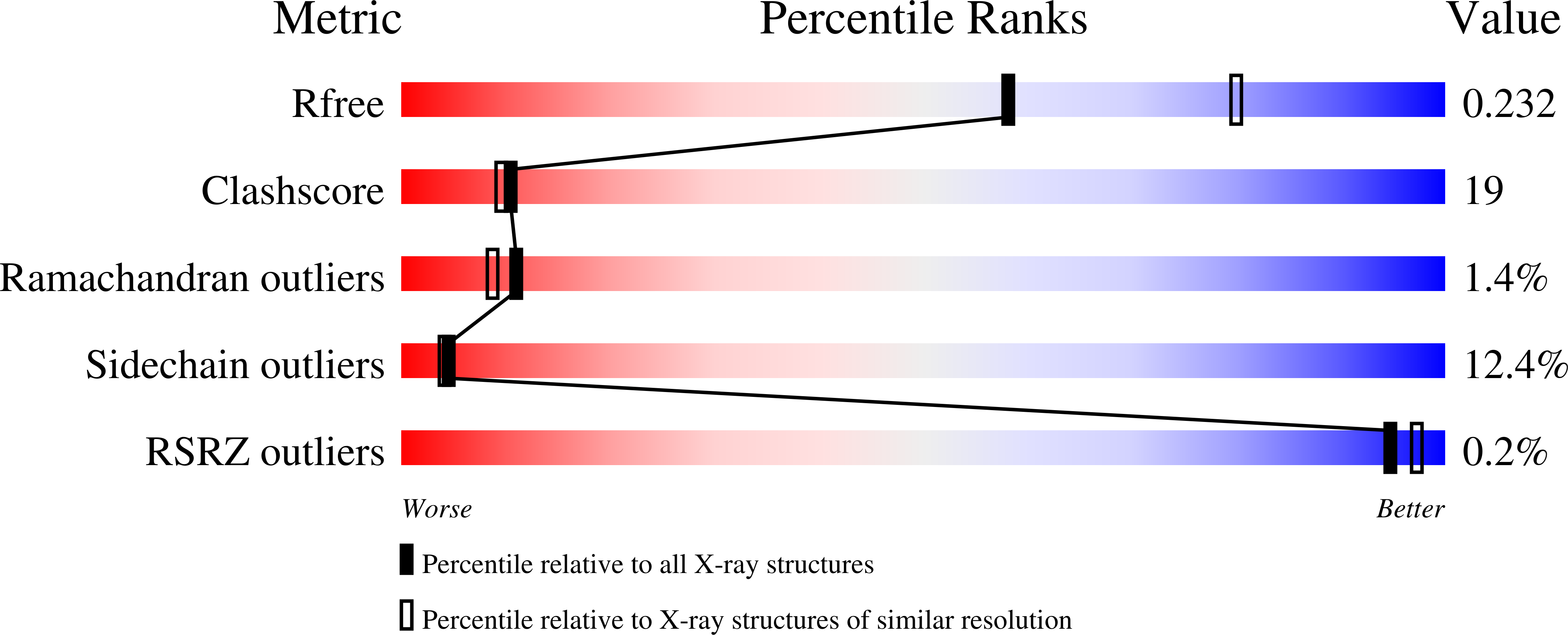
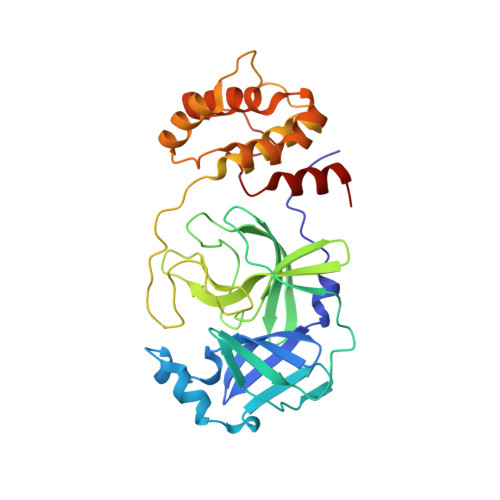
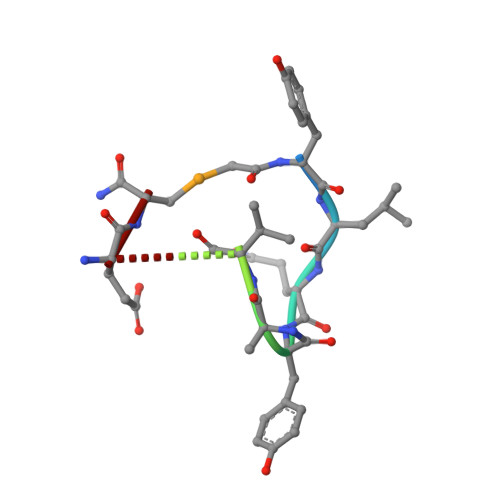
Antiviral cyclic peptides targeting the main protease of SARS-CoV-2.
Johansen-Leete, J., Ullrich, S., Fry, S.E., Frkic, R., Bedding, M.J., Aggarwal, A., Ashhurst, A.S., Ekanayake, K.B., Mahawaththa, M.C., Sasi, V.M., Luedtke, S., Ford, D.J., O'Donoghue, A.J., Passioura, T., Larance, M., Otting, G., Turville, S., Jackson, C.J., Nitsche, C., Payne, R.J.(2022) Chem Sci 13: 3826-3836
- PubMed: 35432913
- DOI: https://doi.org/10.1039/d1sc06750h
- Primary Citation of Related Structures:
7RNW - PubMed Abstract:
Antivirals that specifically target SARS-CoV-2 are needed to control the COVID-19 pandemic. The main protease (M pro ) is essential for SARS-CoV-2 replication and is an attractive target for antiviral development. Here we report the use of the Random nonstandard Peptide Integrated Discovery (RaPID) mRNA display on a chemically cross-linked SARS-CoV-2 M pro dimer, which yielded several high-affinity thioether-linked cyclic peptide inhibitors of the protease. Structural analysis of M pro complexed with a selenoether analogue of the highest-affinity peptide revealed key binding interactions, including glutamine and leucine residues in sites S1 and S2, respectively, and a binding epitope straddling both protein chains in the physiological dimer. Several of these M pro peptide inhibitors possessed antiviral activity against SARS-CoV-2 in vitro with EC 50 values in the low micromolar range. These cyclic peptides serve as a foundation for the development of much needed antivirals that specifically target SARS-CoV-2.
Organizational Affiliation:
School of Chemistry, The University of Sydney Sydney NSW 2006 Australia richard.payne@sydney.edu.au.